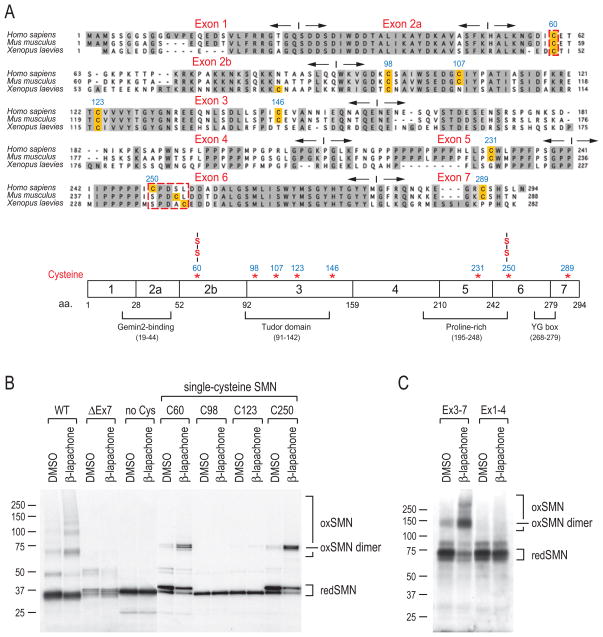
Figure 7. ROS mapping of disulfide-crosslinked cysteines in SMN.
(A) Sequence alignment of human (Homo sapiens), mouse (Mus musculus), and frog (Xenopus laevies) SMN protein sequences. Conserved residues are shaded in gray. Cysteine residues are highlighted. Exons and their boundaries are indicated with opposite-directed arrows. The schematic diagram shows SMN protein domain organization and positions of cysteines. Two cysteines (C60 and C250) that form disulfide bridges are marked (-S-S-).
(B) Human SMN protein (WT, wild type; ΔEx7, exon7 deletion mutant; no Cys, mutation of all 8 cysteines to alanines; C60, C98, C123, and C250, mutation of 7 cysteines to alanines except for cysteine at positions 60, 98, 123, and 250, respectively) were produced by in vitro transcription and translation in the presence of 35S-Met and then treated with 40 μM β-lapachone or DMSO for 1 hour. Samples were mixed with sample buffer without DTT, and then analyzed by SDS-PAGE and autoradiography. Molecular mass markers in kDa are indicated on the left. Protein bands corresponding to monomer SMN (redSMN), disulfide-crosslinked SMN (oxSMN) and SMN dimer (oxSMN dimer) are indicated on the right.
(C) SMN amino terminus deletion (Ex3-7) and carboxyl terminus deletion (Ex1-4) mutants were tested for crosslinking, as described in panel (B). Note that these mutants were constructed in pcDNA3-myc-pyruvate kinase (PK, ~60kD) vector to facilitate detection of otherwise small fragments.