Abstract
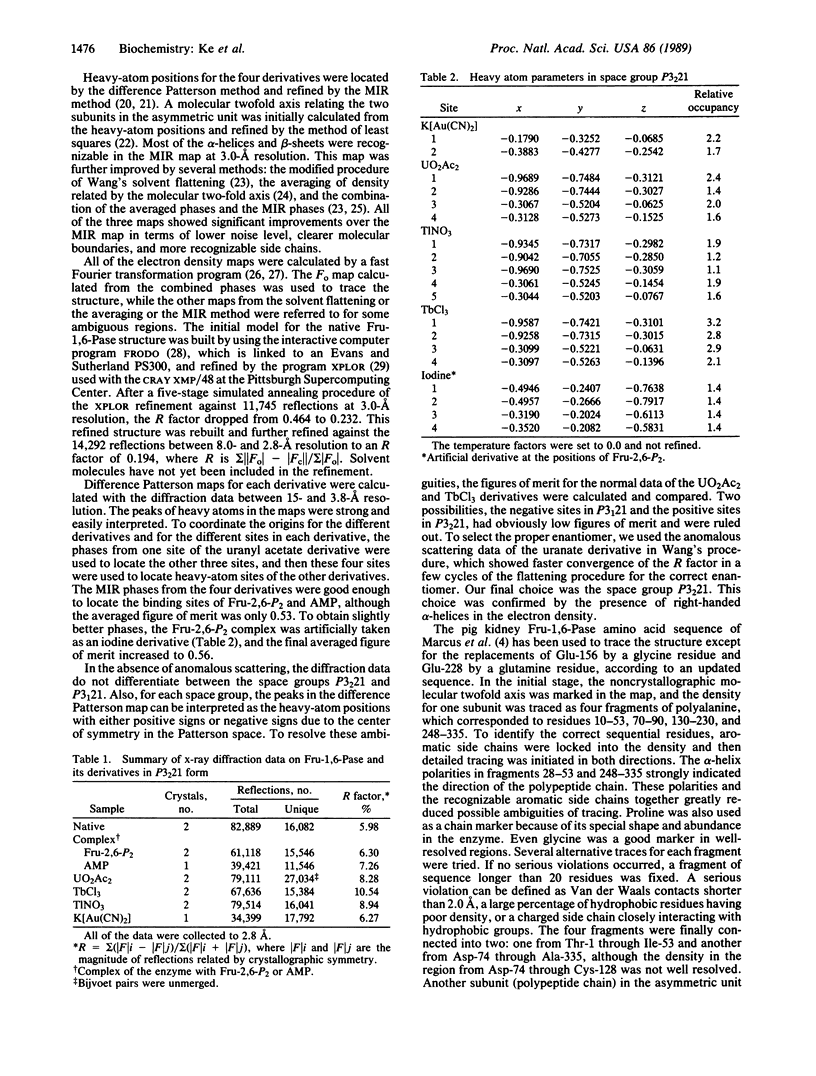
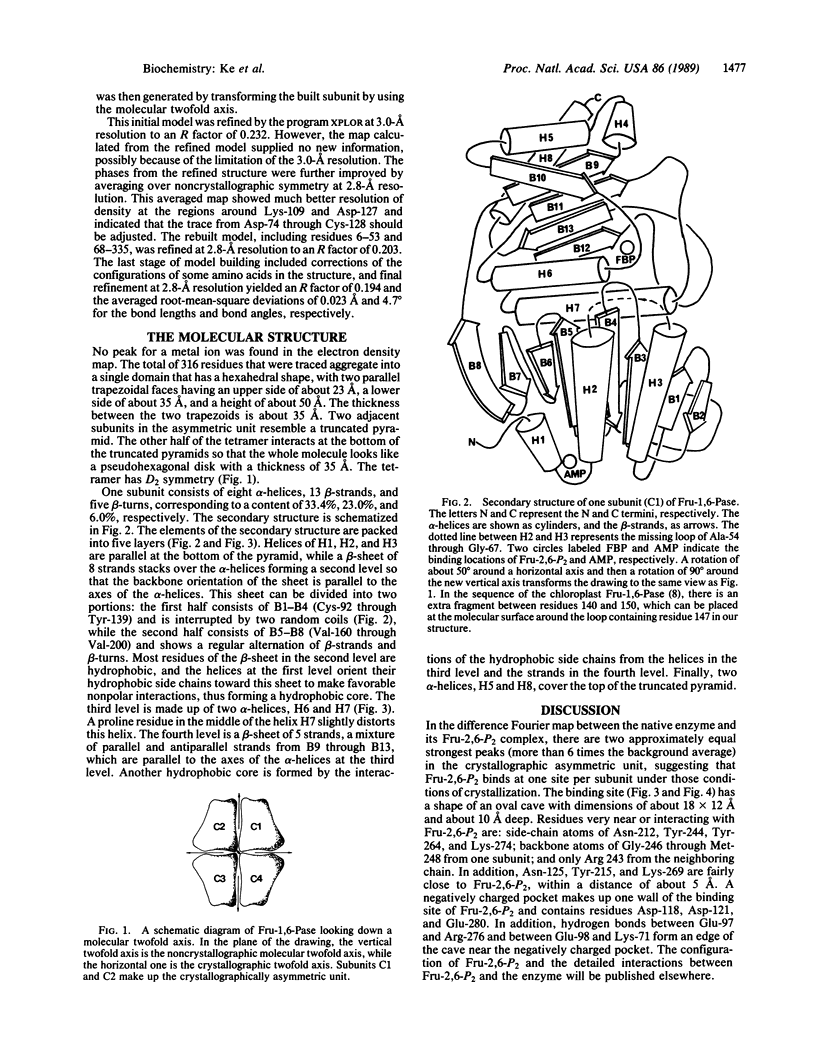
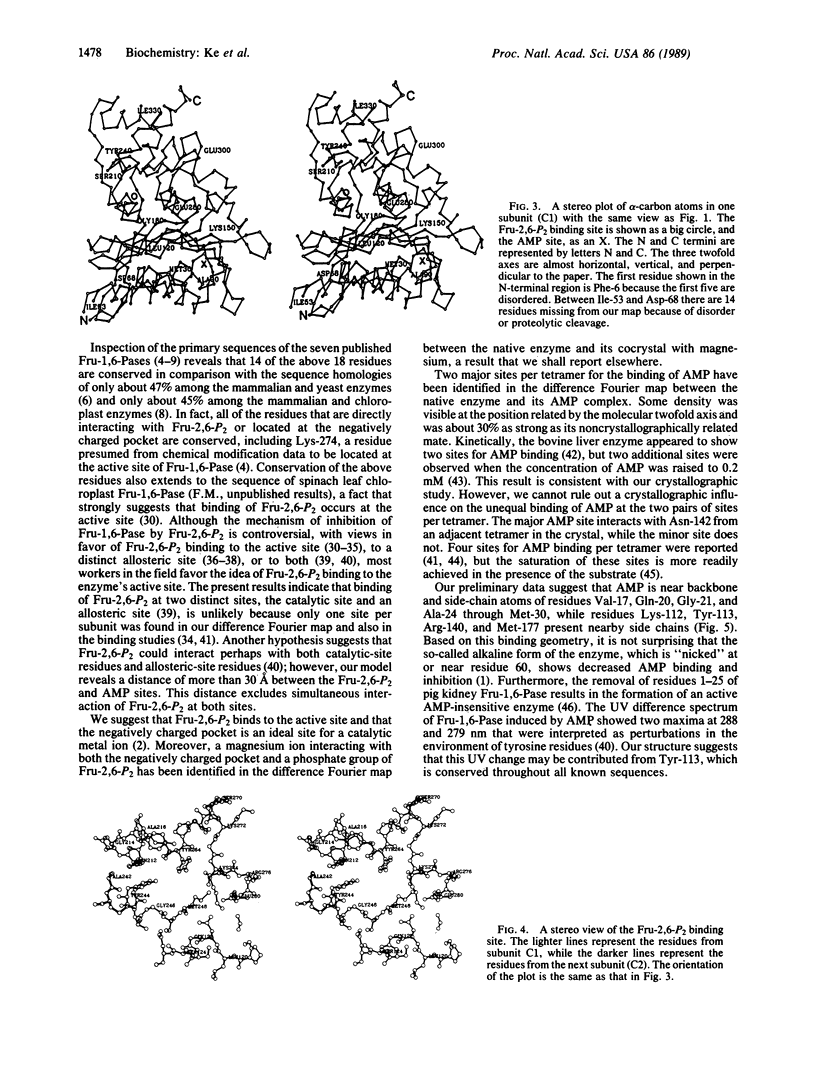
Fructose-1,6-bisphosphatase (D-fructose-1,6-bisphosphate 1-phosphohydrolase, EC 3.1.3.11) from the cortex of pig kidney and its complexes with either fructose 2,6-bisphosphate (Fru-2,6-P2) or adenosine monophosphate (AMP) have been crystallized in the space group P3(2)21. The three-dimensional structure of the native enzyme has been solved at 3.0-A resolution by the multiple isomorphous replacement method and refined at 2.8-A resolution to a crystallographic R factor of 0.194. A total of 316 of 335 residues, omitting disordered regions 1-5 and 54-67, have been built into the monomer, which has average dimensions of about 30 A by 50 A by 35 A. Four monomeric units aggregate into a molecular tetramer with D2 symmetry, which approximates a disk about 35 A thick. Each monomer consists of about 33% alpha-helix, 23% beta-strand, and 6% beta-turn. Four sites for Fru-2,6-P2 and two major sites for AMP binding per tetramer have been identified by difference Fourier techniques. The binding site for Fru-2,6-P2 is shared by two neighboring monomers and consists of side-chain atoms of Asn-212, Tyr-244, Tyr-264, and Lys-274; backbone atoms of Gly-246 through Met-248; and only Arg-243 from the adjacent subunit. In addition, Asn-125, Tyr-215, and Lys-269 are located within a distance of about 5 A of Fru-2,6-P2. A negatively charged pocket near this binding site includes Asp-118, Asp-121, Glu-280, Glu-97, and Glu-98. The AMP binding site is located near Val-17, Gln-20, Gly-21, Ala-24 through Met-30, Lys-112, Tyr-113, Arg-140, and Met-177.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Matthews B. W. Crystallographic data for chicken liver fructose bisphosphatase. J Biol Chem. 1977 Aug 10;252(15):5556–5557. [PubMed] [Google Scholar]
- Arneson R. M., Geller A. M., Byrne W. L. Binding of adenosine 5'-monophosphate to bovine liver fructose 1,6-bisphosphatase. Enzyme. 1979;24(2):132–136. doi: 10.1159/000458641. [DOI] [PubMed] [Google Scholar]
- Benkovic S. J., deMaine M. M. Mechanism of action of fructose 1,6-bisphosphatase. Adv Enzymol Relat Areas Mol Biol. 1982;53:45–82. doi: 10.1002/9780470122983.ch2. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chatterjee T., Reardon I., Heinrikson R. L., Marcus F. Des-1-25-fructose-1,6-bisphosphatase, a nonallosteric derivative produced by trypsin treatment of the native protein. J Biol Chem. 1985 Nov 5;260(25):13553–13559. [PubMed] [Google Scholar]
- Fisher W. K., Thompson E. O. Amino acid sequence studies on sheep liver fructose-bisphosphatase. II. The complete sequence. Aust J Biol Sci. 1983;36(3):235–250. doi: 10.1071/bi9830235. [DOI] [PubMed] [Google Scholar]
- François J., Van Schaftingen E., Hers H. G. On the mechanism of inhibition of neutral liver fructose 1,6-bisphosphatase by fructose 2,6-bisphosphate. Eur J Biochem. 1983 Aug 1;134(2):269–273. doi: 10.1111/j.1432-1033.1983.tb07561.x. [DOI] [PubMed] [Google Scholar]
- Ganson N. J., Fromm H. J. The effect of fructose 2,6-bisphosphate on the reverse reaction kinetics of fructose 1,6-bisphosphatase from bovine liver. Biochem Biophys Res Commun. 1982 Sep 16;108(1):233–239. doi: 10.1016/0006-291x(82)91856-3. [DOI] [PubMed] [Google Scholar]
- Gottschalk M. E., Chatterjee T., Edelstein I., Marcus F. Studies on the mechanism of interaction of fructose 2,6-bisphosphate with fructose-1,6-bisphosphatase. J Biol Chem. 1982 Jul 25;257(14):8016–8020. [PubMed] [Google Scholar]
- Hamilton W. D., Harrison D. A., Dyer T. A. Sequence of the Escherichia coli fructose-1,6-bisphosphatase gene. Nucleic Acids Res. 1988 Sep 12;16(17):8707–8707. doi: 10.1093/nar/16.17.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S., Uyeda K. A binding study of the interaction of beta-D-fructose 2,6-bisphosphate with phosphofructokinase and fructose-1,6-bisphosphatase. J Biol Chem. 1983 Jun 25;258(12):7352–7357. [PubMed] [Google Scholar]
- Kratowich N., Mendicino J. Role of enzyme interactions in the regulation of gluconeogenesis. Modification of the binding properties of fructose 1,6-diphosphatase by adenosine monophosphate, adenosine triphosphate, and fructose 1,6-diphosphate. J Biol Chem. 1974 Sep 10;249(17):5485–5494. [PubMed] [Google Scholar]
- Liu F., Fromm H. J. Relationship between thiol group modification and the binding site for fructose 2,6-bisphosphate on rabbit liver fructose-1,6-bisphosphatase. J Biol Chem. 1988 Jul 15;263(20):10035–10039. [PubMed] [Google Scholar]
- Marcus F., Edelstein I., Reardon I., Heinrikson R. L. Complete amino acid sequence of pig kidney fructose-1,6-bisphosphatase. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7161–7165. doi: 10.1073/pnas.79.23.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus F., Harrsch P. B., Moberly L., Edelstein I., Latshaw S. P. Spinach chloroplast fructose-1,6-bisphosphatase: identification of the subtilisin-sensitive region and of conserved histidines. Biochemistry. 1987 Nov 3;26(22):7029–7035. doi: 10.1021/bi00396a026. [DOI] [PubMed] [Google Scholar]
- McGrane M. M., El-Maghrabi M. R., Pilkis S. J. The interaction of fructose 2,6-bisphosphate and AMP with rat hepatic fructose 1,6-bisphosphatase. J Biol Chem. 1983 Sep 10;258(17):10445–10454. [PubMed] [Google Scholar]
- McPherson A., Burkey D., Stankiewicz P. Crystalline alkaline form fructose-1,6-diphosphatase. A simple purification procedure and preliminary x-ray diffraction analysis. J Biol Chem. 1977 Oct 25;252(20):7031–7034. [PubMed] [Google Scholar]
- Meek D. W., Nimmo H. G. The interaction of fructose 2,6-bisphosphate with an allosteric site of rat liver fructose 1,6-bisphosphatase. FEBS Lett. 1983 Aug 22;160(1-2):105–109. doi: 10.1016/0014-5793(83)80946-6. [DOI] [PubMed] [Google Scholar]
- Muirhead H., Cox J. M., Mazzarella L., Perutz M. F. Structure and function of haemoglobin. 3. A three-dimensional fourier synthesis of human deoxyhaemoglobin at 5.5 Angstrom resolution. J Mol Biol. 1967 Aug 28;28(1):117–156. doi: 10.1016/s0022-2836(67)80082-2. [DOI] [PubMed] [Google Scholar]
- Nimmo H. G., Tipton K. F. The allosteric properties of beef-liver fructose bisphosphatase. Eur J Biochem. 1975 Oct 15;58(2):575–585. doi: 10.1111/j.1432-1033.1975.tb02408.x. [DOI] [PubMed] [Google Scholar]
- Pilkis S. J., El-Maghrabi M. R., McGrane M. M., Pilkis J., Claus T. H. The role of fructose 2,6-bisphosphate in regulation of fructose-1,6-bisphosphatase. J Biol Chem. 1981 Nov 25;256(22):11489–11495. [PubMed] [Google Scholar]
- Pilkis S. J., El-Maghrabi M. R., Pilkis J., Claus T. Inhibition of fructose-1,6-bisphosphatase by fructose 2,6-bisphosphate. J Biol Chem. 1981 Apr 25;256(8):3619–3622. [PubMed] [Google Scholar]
- Pontremoli S., Melloni E., Michetti M., Salamino F., Sparatore B., Horecker B. L. On the mechanism of inhibition of fructose 1,6-bisphosphatase by fructose 2,6-bisphosphate. Arch Biochem Biophys. 1982 Oct 15;218(2):609–613. doi: 10.1016/0003-9861(82)90386-1. [DOI] [PubMed] [Google Scholar]
- Raines C. A., Lloyd J. C., Longstaff M., Bradley D., Dyer T. Chloroplast fructose-1,6-bisphosphatase: the product of a mosaic gene. Nucleic Acids Res. 1988 Aug 25;16(16):7931–7942. doi: 10.1093/nar/16.16.7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Hubert E., Slebe J. C. The reactive cysteine residue of pig kidney fructose 1,6-bisphosphatase is related to a fructose 2,6-bisphosphate allosteric site. Biochem Biophys Res Commun. 1985 Feb 28;127(1):373–379. doi: 10.1016/s0006-291x(85)80169-8. [DOI] [PubMed] [Google Scholar]
- Rogers D. T., Hiller E., Mitsock L., Orr E. Characterization of the gene for fructose-1,6-bisphosphatase from Saccharomyces cerevisiae and Schizosaccharomyces pombe. Sequence, protein homology, and expression during growth on glucose. J Biol Chem. 1988 May 5;263(13):6051–6057. [PubMed] [Google Scholar]
- Sarngadharan M. G., Watanabe A., Pogell B. M. Binding of adenosine 5'-monophosphate and substrate by rabbit liver fructose 1,6-diphosphatase. Biochemistry. 1969 Apr;8(4):1411–1419. doi: 10.1021/bi00832a016. [DOI] [PubMed] [Google Scholar]
- Seaton B. A., Campbell R. L., Petsko G. A., Rose D. R., Edelstein I., Marcus F. Preliminary X-ray crystallographic studies of pig kidney fructose-1,6-bisphosphatase. J Biol Chem. 1984 Jul 25;259(14):8915–8916. [PubMed] [Google Scholar]
- Soloway B., McPherson A. Molecular symmetry of fructose-1,6-diphosphatase by X-ray diffraction analysis. J Biol Chem. 1978 Apr 10;253(7):2461–2462. [PubMed] [Google Scholar]
- TAKETA K., POGELL B. M. ALLOSTERIC INHIBITION OF RAT LIVER FRUCTOSE 1,6-DIPHOSPHATASE BY ADENOSINE 5'-MONOPHOSPHATE. J Biol Chem. 1965 Feb;240:651–662. [PubMed] [Google Scholar]
- Tejwani G. A. Regulation of fructose-bisphosphatase activity. Adv Enzymol Relat Areas Mol Biol. 1983;54:121–194. doi: 10.1002/9780470122990.ch3. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E. Fructose 2,6-bisphosphate. Adv Enzymol Relat Areas Mol Biol. 1987;59:315–395. doi: 10.1002/9780470123058.ch7. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E., Hers H. G. Inhibition of fructose-1,6-bisphosphatase by fructose 2,6-biphosphate. Proc Natl Acad Sci U S A. 1981 May;78(5):2861–2863. doi: 10.1073/pnas.78.5.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- el-Maghrabi M. R., Pilkis J., Marker A. J., Colosia A. D., D'Angelo G., Fraser B. A., Pilkis S. J. cDNA sequence of rat liver fructose-1,6-bisphosphatase and evidence for down-regulation of its mRNA by insulin. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8430–8434. doi: 10.1073/pnas.85.22.8430. [DOI] [PMC free article] [PubMed] [Google Scholar]


