Abstract
The gene encoding the neuronal sortilin-related receptor SORL1 has been claimed to be associated with Alzheimer Disease by independent groups and across various human populations. We evaluated six genetic markers in SORL1 in a sample of 1558 Swedish dementia cases (including 1270 Alzheimer disease cases) and 2179 controls. For both single marker and haplotype-based analyses we found no strong support for SORL1 as a dementia- or AD-risk modifying gene in our sample in isolation, nor did we observe association with AD/dementia-related traits, including CSF β-amyloid1–42, tau levels, or age-at-onset. However, meta-analyses of markers in this study together with previously published studies on SORL1 encompassing in excess of 13,000 individuals does suggest significant association with AD (best OR 1.097; 95% CI 1.038–1.158, p = 0.001). All six markers were significant in meta-analyses and it is notable that they occur in two distinct LD blocks. These data are consistent with either allelic heterogeneity or the existence of as yet untested functional variants and these will be important considerations in further attempts to evaluate the importance of sequence variation in SORL1 with AD risk.
Keywords: SORL1, Alzheimer, meta-analysis, association, Swedish
The gene encoding apolipoprotein E (APOE) remains the only firmly validated genetic risk factor for late-onset Alzheimer disease [1]. One of the more controversial recently claimed candidate genes for Alzheimer is that encoding the neuronal sorting receptor SORL1 [2]. Replication efforts have resulted in a mixture of negative and positive findings, with no present consensus [3–7]. We sought in the present study to attempt replication of previous findings, and have expanded our search for genetic association to include broadly defined dementia as well as AD risk. In addition, given the putative role of SORL1 in amyloid precursor protein (APP) processing, we also sought evidence of genetic association with CSF levels of β-amyloid1–42.
Samples used in the present study were derived primarily from the population-based Swedish Twin Registry [8,9]. An independent non-twin case-control Swedish AD sample was also included (described in detail in ref. 10). In total, DNA was available for 1558 dementia cases and 2179 controls (1270 cases had a possible or probable AD diagnosis). There were 976 men and 1203 women in the control group, and 590 men and 968 women in the dementia group. Average age-at-sampling for controls was 77.7 ± 8.7 (SD) years and age-at-onset for dementia/AD cases was 75.3 ± 8.3 (SD) years. This study was approved by the local ethical review board at Karolinska Institutet.
Markers rs668387, rs689021, rs641120, rs2070045, rs1699102, rs3824968, and rs2282649 were selected for testing in the present study [2]. These were chosen since they include the previously reported most significant markers for AD association and are also the best represented in the literature to facilitate meta-analyses. Genotyping was performed using the Illumina GoldenGate assay system on Illumina BeadStation 500GX equipment, currently housed and implemented at the Uppsala University SNP Technology Platform. Prior to use on the Illumina system, all samples were subjected to Whole Genome Amplification (WGA) using standard kits involving Phi29 DNA polymerase (Amersham). Genotyping failed for marker rs1699102.
CSF samples were obtained in the AD case-control study by lumbar puncture in the L3/L4 or L4/L5 inter-space. Further details of CSF collection can be found elsewhere [11]. CSF Aβ42 was determined using a sandwich enzyme-linked immunosorbent assay (ELISA) (Innotest b-amyloid (1–42), Innogenetics, Ghent, Belgium) constructed to specifically measure Aβ42 [12]. The microtubule-associated protein tau, a CSF marker of neuronal degeneration, was determined using a sandwich ELISA (Innotest hTAU-Ag, Innogenetics, Ghent, Belgium) constructed to measure total tau, i.e., all isoforms of tau irrespective of phosphorylation state [13].
There was no evidence of deviation from Hardy-Weinberg equilibrium (HWE) for any marker. Tests of association between individual markers and both dementia and AD risk were performed using alternating logistic regression (ALR) to account for sibling pair dependencies, allowing both members of the pair to enter in the analyses while accounting for MZ and DZ pair correlation structures [14,15]. Statistical analyses were performed in SAS 9.1 using the GENMOD procedure (SAS Institute, Inc., Raleigh, NC). Haplotypes were estimated after LD block definition in individual blocks using Haploview v4.1 [16]. Tests of genotypes versus quantitative traits (age-at-onset, CSF tau, and CSF Aβ42) were conducted using ANOVA. Logistic regression was used to model potential interaction effects with APOE and gender. Meta-analyses were performed using the “meta”-command in Stata 10.0. Data were combined using both fixed effects and random effects models. Odds ratios presented for meta-analyses are allelic. Heterogeneity was assessed using a Q-test.
Full case-control association results accounting for sibling structure and testing both the broadly defined dementia group and AD cases compared with non-demented controls are shown in table 1. With this study design there was no evidence of association for any of the six tested markers. Notably, the effect of APOE (specifically marker rs429358) is easily detected in these samples (P = 7.8 × 10−23). In table 1 we also show genotype counts when only possible or probable AD cases are tested, where there was again no evidence of association. Haplotype-based tests also failed to provide evidence of genetic association (not shown).
Table 1.
Genetic association of genetic markers in SORL1 with dementia/AD risk and quantitative traits
| Marker | Group | Genotypes | dementia | AD only | AB42 p | Tau p | ||
|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | ||||||
| rs668387 | cases | 279 (219) | 723 (572) | 423 (353) | 0.97(0.87–1.07) | 0.95(0.86–1.06) | 0.36 (639) | 0.76 (655) |
| controls | 444 | 1008 | 595 | p = 0.50 | p = 0.35 | |||
| AA | AG | GG | ||||||
| rs689021 | cases | 274 (213) | 762 (606) | 459 (383) | 0.97(0.88–1.07) | 0.96(0.87–1.06) | 0.52 (674) | 0.75 (691) |
| controls | 437 | 1051 | 649 | p = 0.59 | p = 0.39 | |||
| AA | AG | GG | ||||||
| rs641120 | cases | 282 (221) | 780 (624) | 471 (393) | 0.97(0.88–1.07) | 0.96(0.87–1.06) | 0.45 (704) | 0.75 (721) |
| controls | 448 | 1070 | 666 | p = 0.55 | p = 0.36 | |||
| AA | AC | CC | ||||||
| rs2070045 | cases | 839 (677) | 593 (480) | 101 (80) | 0.99(0.88–1.10) | 0.96(0.86–1.08) | 0.57 (705) | 0.77 (722) |
| controls | 1198 | 841 | 142 | p = 0.82 | p = 0.52 | |||
| AA | AT | TT | ||||||
| rs3824968 | cases | 159 (134) | 702 (559) | 666 (538) | 1.04(0.94–1.15) | 1.08(0.97–1.19) | 0.77 (700) | 0.41 (717) |
| controls | 229 | 967 | 988 | p = 0.43 | p = 0.16 | |||
| AA | AG | GG | ||||||
| rs2282649 | cases | 147 (123) | 687 (554) | 693 (554) | 1.06(0.95–1.17) | 1.10(0.99–1.22) | 0.80 (702) | 0.43 (719) |
| controls | 214 | 924 | 1040 | p = 0.30 | p = 0.078 | |||
Genotype counts are shown for dementia and (AD alone). Odds ratios (95% CI) and significances for case-control association in dementia and AD were determined using alternating logistic regression to account for sibling structure (as described in the main text). Significance for quantitative traits (number of individuals) was performed using ANOVA in unrelated individuals.
All six makers were next tested for possible interaction with APOE e4 carrier status and gender, as well as differences that might be apparent between the twin and non-twin subsets of samples. There was no evidence of interaction effects, nor were the six tested markers significant in any of the strata (not shown).
We evaluated all six markers for possible effects on CSF levels of Aβ42 and tau, as well as age-at-onset for both dementia and AD cases. The results of CSF quantitative trait analyses are shown in table 1 where again we saw no indication of significant effects. CSF trait analyses were performed in cases only. Worth noting is that APOE (rs429358) contributes strongly to variance in CSF Aβ42 in this same sample [17]. There was no evidence of an effect on AAO for any marker (not shown).
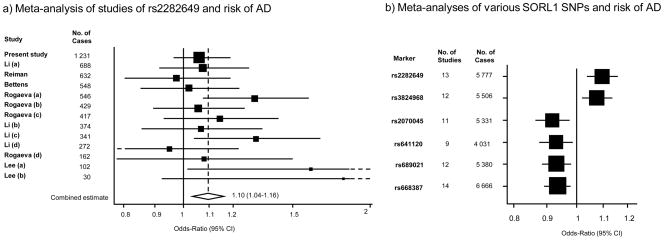
Meta-analyses were conducted by combining the data presented here for the AD population (excluding other forms of dementia) with data available on the Alzgene website (http://www.alzforum.org/res/com/gen/alzgene/), which covers published SORL1 studies to date [18]. Only populations of European ancestry were included. Fixed effects models and random effects models gave similar results for all genotypes. The results from the fixed effects models are given in Figure 1. The corresponding combined estimates as OR (95% CI) based on random effects modeling were as follows for each genotype: rs2282649; 1.10 (1.04–1.17), rs3824968; 1.12 (1.02–1.22), rs2070045; 0.89 (0.82–0.97), rs641120; 0.93 (0.88–0.99), rs689021; 0.93 (0.87–0.99), rs668387; 0.93 (0.88–0.99). Significant heterogeneity was found for rs3824968 (p= 0.01). The combined effects from all six markers were significant (Figure 1b). For the most significant finding, the combined effect size for rs2282649 was OR 1.107 (95% CI 1.049–1.168) in all previous studies and the result from our individual study was 1.099 (CI 0.989–1.221). The current meta-analysis with updated available information gave an effect size estimate for this marker of OR 1.097 (CI 1.038–1.158, p = 0.001). As this was the best evidence across the markers studied here, we present the results of the analysis in Figure 1a. Of note, these six markers occur in two separate LD blocks, with an LD between the two most significant markers in each of the blocks (rs2282649 and rs668387) of around r2 = 0.005. Focusing on rs2282649 we also tested our sample together with previous studies, but excluding the original Rogaeva data sets. The result of this analysis was also significant (OR = 1.077, CI 1.010–1.149, p = 0.023).
Fig. 1.
Meta-analyses of SORL1 genotypes in relation to risk of AD showing a) A literature-based based meta-analysis of individual studies of SNP rs2282649 and b) The combined estimates resulting from individual meta-analyses of six different SNPs, including rs2282649. Studies listed in the meta-analysis for rs22382649 can be found summarized at http://www.alzforum.org/res/com/gen/alzgene/. Results are from fixed effects models. Tests for heterogeneity for each genotype: rs2282649; Q=13.82, p= 0.312, rs3824968; Q=25.60, p=0.013, rs2070045; Q= 18.33, p= 0.05, rs641120; Q=5.88, p= 0.66, rs689021; Q= 14.85, p=0.19, rs668387; Q= 17.21, p=0.19
We noted from a recent genome-wide association study on gene expression in lymphoblast cell lines that sequence variants upstream of the SORL1 promoter were strongly associated with SORL1 transcript levels [19]. We sought replication of that result by examining a genome-wide association study of gene expression in human brain [20]. One of the markers that was highly significant for SORL1 expression in lymphoblasts, rs891437, was also present in the brain study, but was not significant (p = 0.41). This same marker was also present in the genome-wide study included here for meta-analyses [21] (see Figure 1), but was not significantly associated with AD risk (p = 0.96).
In summary, while we have been unable to validate genetic association of SORL1 with AD in our sample in isolation as previously reported [2], we cannot discount the existence of functional polymorphism in SORL1 that may contribute to AD risk, since meta-analysis suggests a genetic effect. In considering emergent genome-wide association studies, simple power estimates based upon a predicted risk effect of OR ~1.1 and minor allele frequency of ~0.3 suggest that sample sizes on the order of 40,000 cases and controls may be required to provide more convincing evidence of association (i.e. survive multiple testing in a genome-wide context). The most significant findings to date were made in the original study by Rogaeva et al [2] and follow on work has successively reduced the predicted effect size estimate(s), making further large-scale studies all the more important. Since markers in each of the two represented LD blocks here show association in meta-analyses, data are consistent with either allelic heterogeneity or the existence of as yet untested sequence variants with greater effect size.
Acknowledgments
We are grateful for generous funding from the US National Institutes of Health (grants AG028555, AG08724, and AG08861) and the Swedish Medical Research Council (grant 2007–2722).
Footnotes
The original publication is available at springerlink.com: www.springerlink.com/index/10.1007/s10048-009-0210-4
References
- 1.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minster RL, DeKosky ST, Kamboh MI. No association of SORL1 SNPs with Alzheimer’s disease. Neurosci Lett. 2008;440:190–192. doi: 10.1016/j.neulet.2008.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 5.Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, et al. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O’Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichtenstein P, De Faire U, Floderus B, Svartengren M, Svedberg P, Pedersen NL. The Swedish Twin Registry: a unique resource for clinical, epidemiological and genetic studies. J Intern Med. 2002;252:184–205. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen NL, Lichtenstein P, Svedberg P. The Swedish Twin Registry in the third millennium. Twin Res. 2002;5:427–432. doi: 10.1375/136905202320906219. [DOI] [PubMed] [Google Scholar]
- 10.Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, Brookes AJ, Blennow K, Kehoe PG, Prince JA. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- 11.Andreasen N, Minthon L, Clarberg A, Davidsson P, Gottfries J, Vanmechelen E, Vanderstichele H, Winblad B, Blennow K. Sensitivity, specificity, and stability of CSF-tau in AD in a community-based patient sample. Neurology. 1999;53:1488–1494. doi: 10.1212/wnl.53.7.1488. [DOI] [PubMed] [Google Scholar]
- 12.Vanmechelen E, Vanderstichele H. Towards an earlier diagnosis of Alzheimer’s disease. J Biotechnol. 1998;66:229–231. doi: 10.1016/s0168-1656(98)00169-2. [DOI] [PubMed] [Google Scholar]
- 13.Blennow K, Wallin A, Agren H, Spenger C, Siegfried J, Vanmechelen E. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol Chem Neuropathol. 1995;26:231–245. doi: 10.1007/BF02815140. [DOI] [PubMed] [Google Scholar]
- 14.Carey V, Zeger SL, Diggle P. Modelling multivariate binary data with alternating logistic regressions. Biometrika. 1993;80:517–526. [Google Scholar]
- 15.Jansson M, Gatz M, Berg S, Johansson B, Malmberg B, McClearn GE, Schalling M, Pedersen NL. Association between depressed mood in the elderly and a 5-HTR2A gene variant. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:79–84. doi: 10.1002/ajmg.b.20016. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Prince JA, Zetterberg H, Andreasen N, Marcusson J, Blennow K. APOE epsilon4 allele is associated with reduced cerebrospinal fluid levels of Abeta42. Neurology. 2004;62:2116–2118. doi: 10.1212/01.wnl.0000128088.08695.05. [DOI] [PubMed] [Google Scholar]
- 18.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 19.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 20.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 21.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, et al. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]