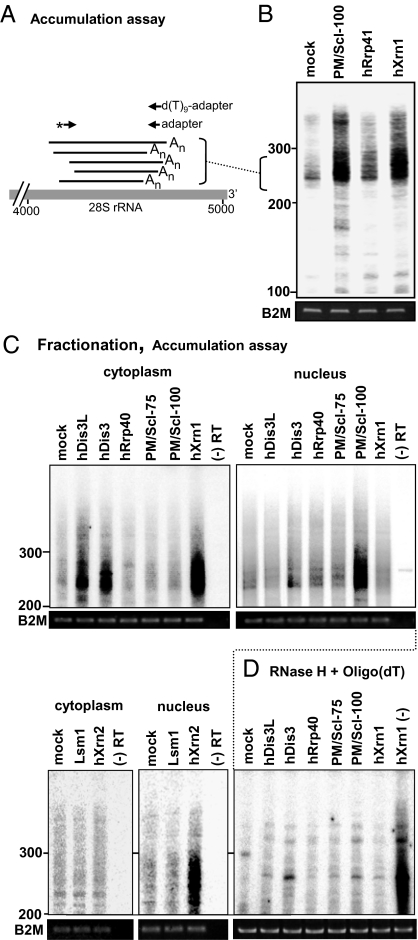
Fig. 4.
Adenylated 28S rRNA degradation intermediates significantly accumulated in cells upon RNAi silencing of major exonucleases. (A) Schematic presentation of PCR-labeling assay to detect accumulation of adenylated, truncated 28S rRNA molecules after exonuclease KD. Briefly, following oligo(dT) RT of RNA from different KD cells, PCR with [32P]-labeled gene-specific primer (asterisk; same as used to isolate molecules as shown in Fig 1B) and adapter oligo was performed. (B) RNA purified from the mock and KD HeLa cells was subjected to this analysis to assess the accumulation of polyadenylated, fragmented 28S rRNA. The migration of DNA markers is shown to the left (nucleotide numbers). (C) Cytoplasmic or nuclear RNA from cells in which the indicated proteins were silenced by RNAi, was subjected to oligo(dT) RT-PCR labeling. A control, with reverse transcriptase omitted from the reaction, is shown in the far right lane of each panel. (D) To verify that the PCR products originated from polyadenylated transcripts, a control was performed in which the poly(A) tails of the cytoplasmic fractions were removed by incubation with oligo(dT) and RNase H before oligo(dT)-primed RT-PCR labeling. This resulted in reduction of amplification signals to background levels, as compared with the hXrn1 sample that was treated with RNase H but without oligo(dT) [lane labeled hXrn1 (-)].