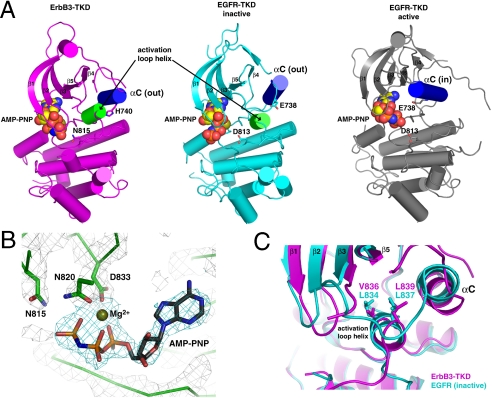
Fig. 3.
Crystal structure of ErbB3-TKD665–1001. (A) Cartoon representations are shown for ErbB3-TKD665–1001 (Left: Magenta), inactive EGFR-TKD (Middle: Cyan), and active EGFR-TKD (Right: Gray). EGFR-TKD structures are from PDB codes 2GS7 (5) and 2ITX (48). All structures have bound AMP-PNP and Mg2+ (shown in spheres). Helix αC is colored blue in each structure and is denoted as out (ErbB3 and inactive EGFR) or in (active EGFR). The short activation loop helix in ErbB3 and inactive EGFR is colored green. H740 in helix αC of ErbB3 and the similarly located E738 in EGFR are shown as sticks, as are ErbB3 N815 and EGFR D813. β-strands in the N lobe are labeled. (B) Close-up view of Mg2+-AMP-PNP in the ErbB3-TKD665–1001 active site, with electron density shown as a 2Fo - Fc map calculated at 1.8σ by using phases from a model with no nucleotide. Coordination of bound Mg2+ by the N820 and D833 side chains (from the DFG motif) and the AMP-PNP α- and β-phosphates is depicted. N815 (close to the γ-phosphate) is also marked. (C) Close-up of the short activation loop helix seen in ErbB3-TKD665–1001 (Magenta) and inactive EGFR-TKD (Cyan), with the two kinase domains superimposed. Mutations at L834 or L837 in EGFR activate the receptor in NSCLC. These residues overlay with V836 and L839 in ErbB3-TKD665–1001 and interact with the hydrophobic pocket that contributes to stabilization of helix αC in the out position. A V836A mutation inhibits ErbB3-TKD activity.