Fig. 4.
Schematic free energy landscape for DNA sequence recognition by E2C. (A) Illustration of free energy as a function of the similarity to the native complex (Qn, X-Axis) and to Ilate (Qt, Y-Axis), at conditions where both the native complex and the unbound reagents are populated. Low free energy regions are in Blue, high free energy regions in Red. The free energy minimum for the unbound reagents (U) is at the lower left corner of the plot, the native complex (N) is at the lower right corner and Ilate at the upper left corner. The two-state kinetic route takes place along the x-axis with the formation of native-like contacts. The multistate kinetic route takes place first along the y-axis with the formation of nonnative contacts in the encounter complex and Ilate, and then along the diagonal with the simultaneous disruption of nonnative contacts and formation of native-like ones to reach the final complex. (B) Double funnel projection of the free energy landscape (36). The width of the plot represents the conformational entropy of the E2C:DNA system, and its depth represents the energy of interaction between the two molecules. The top of the picture corresponds to non-interacting E2C and DNA, and the two funnels group native-like and nonnative protein-DNA interactions. The two-state kinetic route takes place in the minimally frustrated native funnel (Right), centered on the free energy minimum for the native complex. The multistate kinetic route maps to a frustrated nonnative funnel (Left) that is centered on Ilate. Conversion of this intermediate into the global minimum requires the E2C:DNA complex to switch to the native funnel over 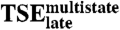 . Dissociation of the native complex may take place over TSE2-state or
. Dissociation of the native complex may take place over TSE2-state or 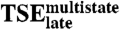 .
.

