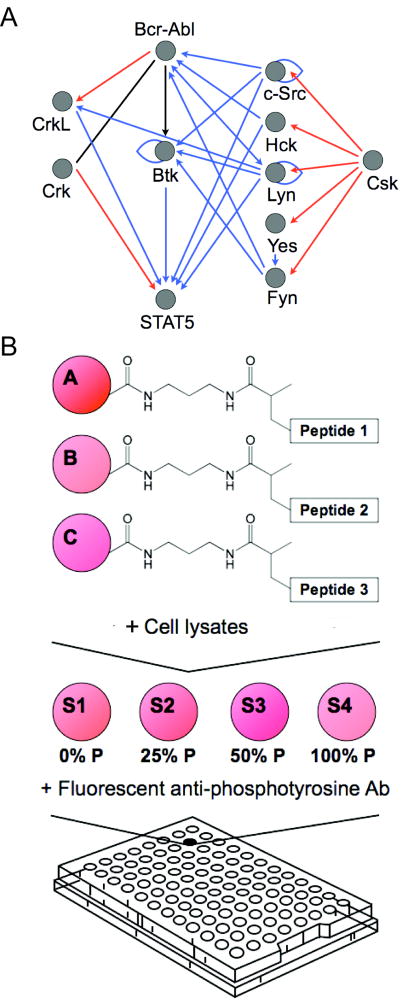
Figure 1.
A, a simplified network of intracellular Bcr-Abl kinase signaling, showing only direct interactions between the kinase Bcr-Abl, adapter proteins Crk and CrkL, the transcription factor Stat5, the kinase Btk, and select members of the Src family of kinases, c-Src, Hck, Lyn, Yes, Fyn, and Csk. These proteins have been shown to play a role in the initiation, progression, or maintenance of CML though either up-regulation, over-expression, or by interaction with the inhibitors imatinib and dasatinib. The functional network was constructed using the MetaCore™ (GeneGo, St. Joseph, MI) database of reported interactions manually curated from the literature. Blue arrows represent protein binding or substrate phosphorylation that leads to activation, while red arrows represent interactions that lead to inhibition. A black line represents interactions that have been activating or inhibiting under different conditions. B, experimental scheme. To monitor tyrosine kinase activity in a signaling network, we covalently immobilized peptide substrates on Luminex beads via an acrylamide linker. Beads bearing different peptide substrates were combined in a single well and reacted with cell lysates. A set of four synthetically phosphorylated internal standards were added to each well in a 96-well array following the termination of kinase reactions and prior to labeling with fluorescent antibodies against phospho-tyrosine. Non-linear regression from well-specific internal standard curves was used to calculate the percentage of substrate phosphorylation. This allowed for accurate comparisons between wells in an array and between arrays.