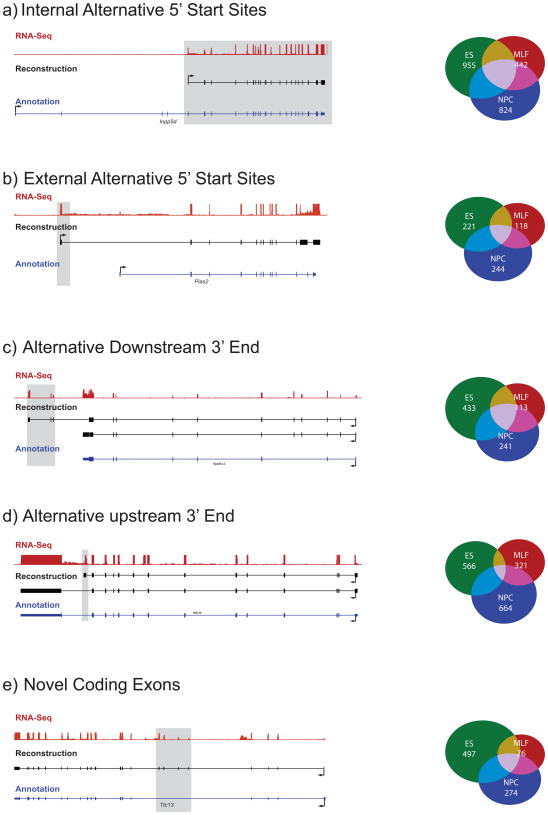
Figure 3. Alternative 5′ ends, 3′ ends and novel coding exons in transcripts reconstructed by Scripture.
Shown are representative examples (tracks, left) and summary counts (Venn diagrams, right) of five categories of variations discovered in Scripture transcripts compared to the known annotations. In each representative example, shown is the coverage by RNA-Seq reads (top track, red), the reconstructed annotation (middle track, black), and the known annotation (bottom track, blue). The novel regions in the reconstruction are marked by gray shading. In each proportional Venn diagram we show the number of transcripts in this class in each cell type (ESC – green, NPC – blue, MLF – red) and their overlap. (a) Internal alternative 5′ start; (b) External alternative 5′ start; (c) Alternative downstream 3′ end (extended termination); (d) Alternative upstream 3′ end (early termination); (e) Novel coding exons.