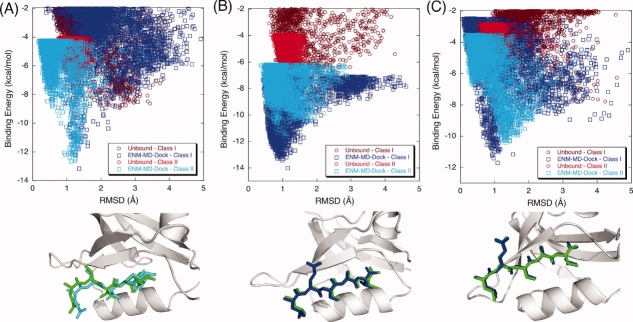
Figure 4.
The binding energy score versus RMSD of the docked complex for PICK1. Wild-type PICK1 (A) prefers Class II type of peptide sequence while K25E mutation (B) and K83H mutation (C) on PICK1 alter its binding specificity to Class I type of peptide sequences. Docking Class I and Class II peptides to the unbound conformation of wild and mutant PICK1 does not show the change in selectivity upon mutation [brown (Class I) and red (Class II) dots in the plots]. However, when REMD-ENM conformations are used, wild type has a higher affinity for Class II type peptide binding [cyan on (A)] whereas both mutants prefer Class I type of peptide [blue on (B) and (C)]. The corresponding lowest energy structures of PDZ-peptide complexes are represented as ribbon diagrams along with experimental peptide position. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]