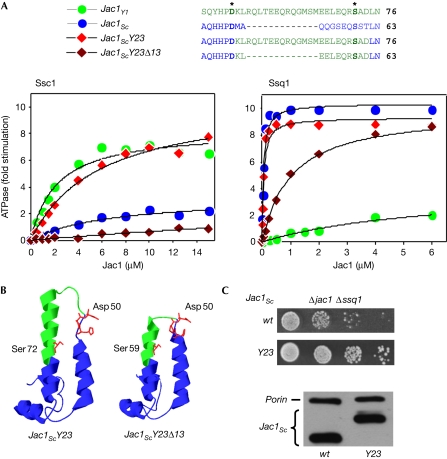
Figure 2.
J-domain structure determines partnership with Ssq1 and Ssc1. (A) Stimulation of the ATPase activity of Ssc1 and Ssq1 from Saccharomyces cerevisiae by chimaeral Jac1ScY23 and Jac1ScY23Δ13 proteins in the presence of Isu1Sc. Data from Fig 1A of stimulation by Jac1Sc and Jac1Yl is included for comparison. The curves represent the best fit of the data to a Michaelis–Menten hyperbolic equation (supplementary Table S1 online). (B) Amino-acid sequence and structural model of Jac1ScY23 and Jac1ScY23Δ13. Green and blue represent sequences derived from Yarrowia lipolytica and S. cerevisiae, respectively. (C) Overexpression of Jac1ScY23 suppresses the growth defect of Δjac1 Δssq1. Top panels: 10-fold serial dilutions (starting with approximately 104 cells) of Δjac1 Δssq1 cells expressing plasmid-borne wild-type Jac1Sc (wt) or Jac1ScY23 (Y23) were plated on a rich glucose medium and incubated at 30°C for 3 days. Bottom panel: cell lysate from the indicated strains separated by electrophoresis and subjected to immunoblot analysis using antibodies specific for Jac1Sc and, as a loading control, porin. Predicted molecular weight: Jac1Sc, 21.6 kDa; Jac1ScY23, 23.3 kDa. Sc, Saccharomyces cerevisiae; Yl, Yarrowia lipolytica.