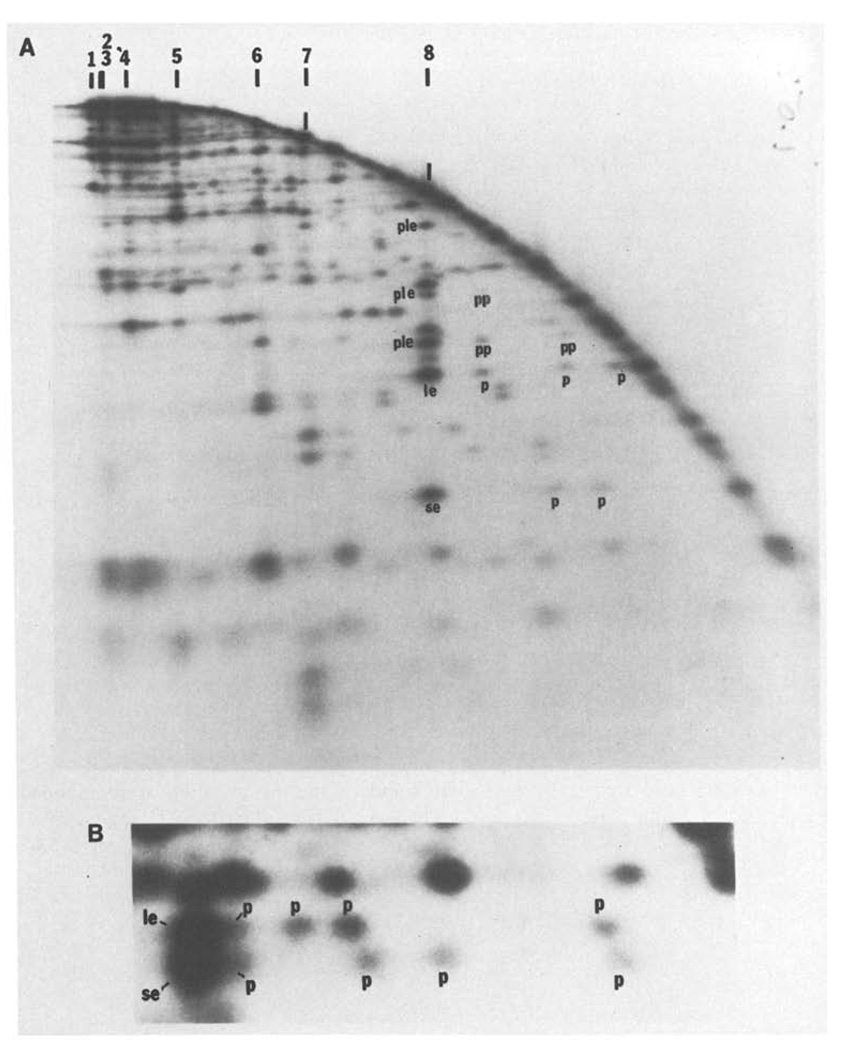
Fig. 5.
A and B. An example of two-dimensional analysis of partial digests. T4 DNA was digested with SalI, end-labeled and then redigested with a low level of HindIII to generate a partial digest. This partial digest was separated on a 0.5% agarose first dimensional gel. The separated fragments were then digested further with HindIII and electrophoresed through a second dimensional 1.5% agarose gel. Panel A shows an autoradiogram of the entire separation. The arc dominating the pattern is generated by those fragments which were not recut prior to electrophoresis in the second dimension. A fraction of each SalI fragment remains intact during the first partial HindIII digestion and in the first dimension migrates to a position characteristic of the intact SalI fragment. These positions are indicated along the top of the figure. When recut with HindIII and electrophoresed in the second dimension, two terminal labeled SalI HindIII fragments will be generated from each of these SalI fragments. For example, SalI 8 generates a small end fragment (se) and a large end fragment (le) whose positions are indicated in panel A. Furthermore, the first partial HindIII digestion will have generated some labeled partials extending from one end to internal HindIII sites. These partials will migrate faster in the first dimension than the parent SalI fragment. When redigested with HindIII and anlyzed in the second dimension, each of these partials will generate one of the same labeled end fragments produced from the intact SalI fragment (these are labeld p). Thus, SalI 8 generates three detectable partials extending from the end with the large end fragment, le, and two detectable partials extending from the other end, se (one of the se partials was not detected on this gel). The size of each partial gives the distance of a HindIII site from the terminus. Although not intended, the “in situ” digestion prior to running this particular second dimension was also a partial digestion. The resulting partials seen in the second dimension complicate the pattern but can be interpreted. For example, the intact SalI 8 fragment generates in addition to the end fragments a set of partials which, extend to the end defined as le (ple). The larger partials produced by the first partial digestion can also produce lower molecular weight partials in the second dimension. These partials of partials are labeled pp. Note that the molecular weight of fragments extending from, for example, the le end of SalI 8 can be determined from the positions of the partials, p, in the horizontal direction or from the positions of the partials produced in the second dimension, ple, or from the positions of partials of partials, pp. All of these determinations agree. Furthermore, the same map can be obtained from an analysis of fragments extended to the other end, se, of SalI 8. Again the data agrees. - Panel B shows an enlargement of a region of the separation in A, and includes both the large end fragment, le, and the small end fragment, se, generated from SalI 7. The area enlarged in panel B is from the low molecular weight region of the second dimension gel and the equivalent area cannot be readily seen in panel A because it depicts a lower level of autoradiographic exposure. For both le and se, three partials p are clearly resolved, a fourth is only partially resolved and a fifth which is nearly as large as SalI 7 cannot be detected. The size of the four resolved partials determines the positions of 4 HindIII sites and the size of the end fragment defines a fifth. Thus, the size of the small end fragment (se), 0.29 kb, shows that there must be a HindIII site at this distance from the end. The smallest partial, 1 kb, defines the position of the next site and so on. Similarly the positions of the HindIII sites can be read from the other direction by examining le. Independent maps are generated by reading from the two ends and these are consistent with each other. In this case the two maps overlapped for 4 HindIII sites and the map generated by se defines an additional site not seen in the map generated by the other end, le. Similarly, the map generated by le detected one site not seen in the map read from se. Thus, there are six HindIII sites. Although two partials went undetected, the redundancy of the data allows construction of a complete map, and as above this can be confirmed by measurement of partials in the second dimension. It should be noted that the two partials which were not detected would according to their predicted size not be resolvable from intact SalI 7. For large fragments, all the partials are not resolved, but a map of the sites near each end can be constructed. This particular gel could be used to determine all the HindIII sites in SalI 5, SalI 6, SalI 7 and SalI 8 and some of the sites in SalI 1 and SalI 4