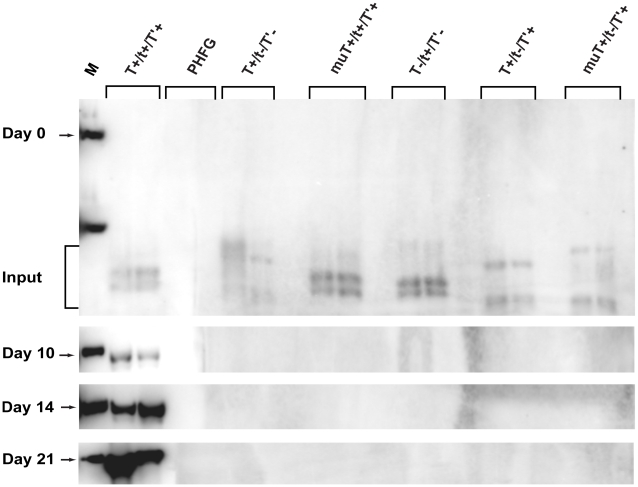
Figure 5. Replication efficiencies of DNA constructs expressing combinations of JCV early proteins.
PHFG cells in 60 mm plates were transfected in duplicate with 400 ng of wild type (T+/t+/T'+) or mutant (T+/t−/T'−, muT+/t+/T'+, T−/t+/T'−, T+/t−/T'+, or muT+/t−/T'+) JCV genomes. At each time point, viral DNA was extracted by the Hirt procedure [39], purified, digested with DpnI and EcoRI, electrophoresed on a 0.8% agarose gel, transferred to nitrocellulose membranes and hybridized to a [P32]dCTP-labeled JCV DNA probe. Bands were visualized with a Typhoon phosphorimager and quantitated with ImageQuant software. The marker (M) shown in the first lane of each blot is 1 ng of linear JCV DNA (5130 bp). The duplicate, independent samples representing each DNA construct were analyzed, and the position of Dpn1-resistent replicating genomes on the Southern blots is denoted by an arrow at days 10, 14 and 21 p.t. Dpn1- and EcoRI-sensitive input DNAs are noted at the day 0 time point. Hirt extracts of uninfected PHFG cells (PHFG) were subjected to the same Dpn1 assay.