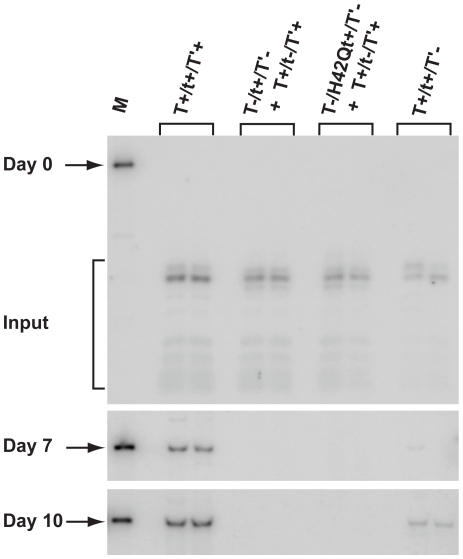
Figure 7. JCV tAg fails to complement replication of a tAg-deficient JCV genome in trans.
PHFG cells in 60 mm plates were co-transfected in duplicate with 400 ng of a tAg-deficient (T+/t−/T'+) JCV genome and 400 ng of a JCV DNA construct expressing either wild type tAg (T−/t+/T′−) or a J domain mutant tAg (T−/H42Qt+/T′−) under the control of the JCV promoter-enhancer. Cells were also transfected with 400 ng of either a T′-deficient (T+/t+/T'−) JCV genome or a positive replication control (T+/t+/T'+). Duplicate, independent samples representing each DNA construct were extracted by the method of Hirt [39] on days 0, 7, and 10 p.t. and analyzed using the Dpn1 assay as described in the legend to Figure 5. The marker shown in the first lane of each blot is 1 ng of linear JCV DNA (5130 bp), and the position of Dpn1-resistent replicating genomes is denoted by an arrow at days 7 and 10 p.t. Dpn1- and EcoRI-sensitive input DNAs are noted at the day 0 time point.