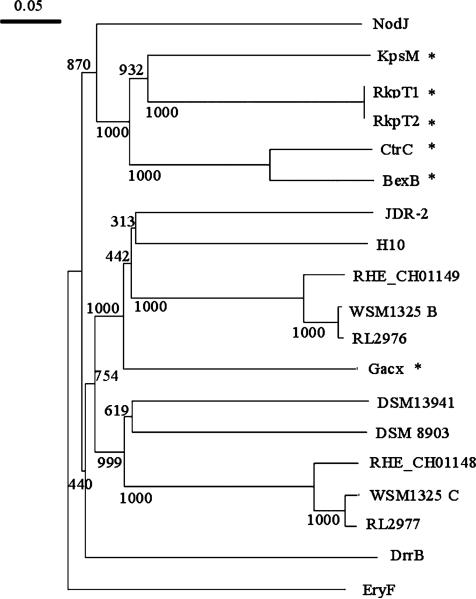
Fig. 2.
Neighbor-joining tree of RL2976 and RL2977 with the transmembrane components of well-characterized ABC-2 transporters and DUF990 proteins. Bootstrap analysis was performed with 1000 replicates, and values (out of 1000) are as displayed on tree. RHE_CH01148, Rhizobium etli CFN 42 (YP_468682.1); RL2977, Rhizobium leguminosarum bv. viciae 3841 (YP_768561.1); DrrB, Mycobacterium avium ssp. paratuberculosis K-10 (NP_960171.1); NodJ, R. leguminosarum bv. viciae 3841 (YP_770467.1); CtrC, Neisseria meningitidis Z2491 (NP_283043.1); BexB, Haemophilus influenzae (P22235); KpsM, Escherichia coli (AAC38078.1); RHE_CH01149, R. etli CFN 42 (YP_468683.1); RL2976, R. leguminosarum bv. viciae 3841 (YP_768560.1); RkpT1, Sinorhizobium meliloti 1021 (NP_437116); RkpT2, S. meliloti 1021 (NP_437102); JDR-2, Paenibacillus sp. JDR-2 (ZP_02846839); H10, Clostridium cellulolyticum H10 (ZP_01574145.1); WSM1325C, R. leguminosarum bv. trifolii WSM1325 (ZP_02293733); DSM 13941, Roseiflexus castenholzii DSM 13941 (YP_001431939); DSM 8903, Caldicellulosiruptor saccharolyticus DSM 8903 (YP_001179931.1); WSM1325B, R. leguminosarum bv. trifolii WSM1325 (ZP_02293734); EryF, R. leguminosarum bv. viciae 3841 (YP_764713); GacX, Streptomyces glaucescens (CAL64857.1). *Represents proteins where experimental data exists on their cellular function.