Abstract
We use elastic neutron scattering to demonstrate that a sharp increase in the mean-squared atomic displacements, commonly observed in hydrated proteins above 200 K and often referred to as the dynamical transition, is present in the hydrated state of both native and denatured lysozyme. A direct comparison of the native and denatured protein thus confirms that the presence of the transition in the mean-squared atomic displacements is not specific to biologically functional molecules.
Keywords: Biomolecules, Dynamics, Neutron scattering
Introduction
The temperature dependence of dynamics of all hydrated biomolecules, such as proteins, DNA, and RNA, exhibits an apparent change at 200–230 K, often referred to as a dynamical transition. In a typical measurement, the transition manifests itself as a sharp increase in the mean-squared atomic displacements, < x2 >, above 200–230 K [1–12]. The origin of the transition has been extensively debated [13–28], and there have been numerous attempts to relate it to the onset of biological activity. Such a connection might seem intuitive because the onset of biological activity and the transition in the mean-squared displacements are both dependent on temperature and hydration level. This hypothesis has become well known [29–32], even though contra arguments [13, 33–35] have been presented.
In this work, we use elastic neutron scattering to demonstrate that the transition in the atomic mean-squared displacements is observed in the hydrated state of both native and irreversibly denatured protein lysozyme, which clearly confirms the view that this transition is not specific to functional biological molecules. This observation suggests that the onset of bioactivity in functional biological molecules does not depend solely on the transition in the mean-squared displacements. The relationship between the onset of anharmonic dynamics and biological activity must be, at best, indirect, as the anharmonicity is apparent even in the intrinsically inactive, irreversibly denatured protein.
Material and methods
The labile hydrogen atoms in chicken egg white lysozyme (Sigma Aldrich L4919; 98% purity) were exchanged for deuterium atoms by dissolving in D2O followed by lyophilization. This process was repeated at least twice for each sample. The denatured lysozyme sample was prepared by dissolving the sample at a concentration of 50 mg/ml in 40 mM NaOD and heating it to 353 K for 30 min followed by lyophilization. Subsequently, the denatured sample underwent one additional D2O exchange step. The samples were hydrated using isopiestic conditions by incubation in a sealed container containing 99.9% D2O. The level of hydration was controlled by varying the incubation time. The final hydration levels of the native and denatured lysozyme were 33.6% and 35.5%, respectively. Circular dichroism spectra were recorded on a Jasco 810 CD spectropolarimeter from 190–240 nm at 298 K. Neutron-scattering measurements were performed on the backscattering spectrometer BASIS (Spallation Neutron Source, Oak Ridge National Laboratory, USA) [36] operated in the regime of elastic intensity scan [37]. The scattering momentum transfer range of 0.5 Å − 1 < Q < 1.7 Å − 1 was used. The Q-averaged energy resolution in the experiment was 3.5 μeV; thus, motions on the time scale of about 0.4 ns and faster were probed. Following cooling down to 20 K, the elastic scattering signal was collected at a heating rate of 1 K/min.
Results and discussion
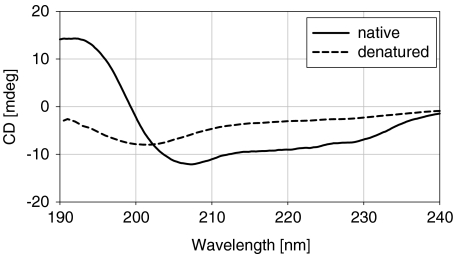
As described in Section 1, the overall aim of this work was to investigate if biological activity in functional molecules is a prerequisite for the dynamical transition that is observed at 200–230 K. Lysozyme was chosen for this study because it has been extensively studied and has well-characterized properties. The protein was denatured under acid and alkali conditions by heating to 353 K, followed by cooling to room temperature. The extent of denaturation of the protein was assessed by circular dichroism spectropolarimetry. After denaturation of lysozyme under acidic conditions, natively folded protein could be detected in the soluble fraction of the sample. Conversely, under alkali conditions, no native lysozyme was detected. A comparison of native and alkali-denatured lysozyme is shown in Fig. 1 and demonstrates clearly that the native conformation of the protein had been disrupted. A deconvolution algorithm [38–40] was used to quantitatively estimate the fraction of each type of secondary structure present in lysozyme in its native and denatured state (Table 1). The α-helical content of the denatured protein was greatly reduced while the β-sheet content and amount of unordered polypeptide had increased, compared to its native counterpart (Table 1). This analysis gives good confidence that the protein is in a non-native conformation after incubation in NaOD at 353 K.
Fig. 1.
Circular dichroism analysis of native and alkali-denatured lysozyme
Table 1.
Fractions of each type of secondary structure present in native and denatured lysozyme
| H(r) | H(d) | S(r) | S(d) | Trn | Unrd | |
|---|---|---|---|---|---|---|
| Native lysozyme | 0.18 | 0.17 | 0.06 | 0.06 | 0.22 | 0.31 |
| Denatured lysozyme | 0.03 | 0.04 | 0.12 | 0.07 | 0.21 | 0.62 |
H(r) regular α-helix, H(d) distorted α-helix, S(r) regular β-sheet, S(d) distorted β-sheet, Trn turn, Unrd unordered
The scattering intensity from the samples is dominated by the non-exchangeable H atoms in the protein; the contribution from the D atoms in the protein and hydration water is weaker, though not negligible. The temperature dependence of the mean-squared displacements, averaged over all the atoms in the samples, was estimated using a Gaussian approximation for the elastic intensity [41]:
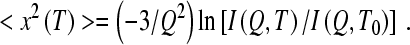 |
1 |
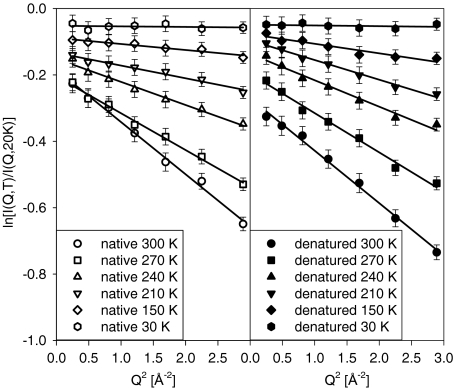
Here T0 = 20 K is the lowest temperature point of the elastic scans, at which most of the atomic motions are suppressed. As one can see in Fig. 2, the Gaussian approximation appears to remain satisfactory through the entire Q range of our experiment, even at the highest measured temperature of 300 K. Even though this approximation is typically used for Q values not exceeding 1 Å − 1, it has been suggested to remain applicable to much higher Q values [42].
Fig. 2.
Fits of the Q-dependence of the elastic neutron scattering intensities from hydrated lysozyme normalized to the 20 K data
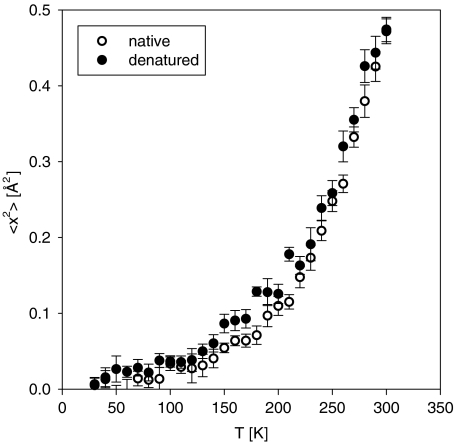
The temperature dependence of the mean-squared displacements for the native lysozyme presented in Fig. 3 is also similar to the previously reported data [7, 10, 12]. Native lysozyme, which possesses a large number of methyl groups, exhibits some anharmonicity (previously attributed to methyl group rotations [7, 10]) above 100 K, followed by the transition and rapidly increasing amplitudes of the atomic displacements above 200 K. Remarkably, denatured lysozyme exhibits a temperature dependence qualitatively similar to that of the native lysozyme, with the transition in the mean-squared atomic displacements clearly present in both states. The difference between the values of the mean-squared displacements for native and denatured lysozyme is rather small, possibly because the large contribution from the methyl groups masks any change of atomic motions that might result from denaturation. The earlier measurement [43] carried out at ambient temperature could not quantify the difference in the mean-squared displacements between native and denatured yeast phosphoglycerate kinase, yielding the same value of (0.42 ± 0.03) Å2 within the experimental error. Nevertheless, in the current temperature-dependent measurement the displacements appear to be systematically larger in the denatured sample. This can be intuitively understood as a result of unfolding of the initially much more compact native protein into a more random-coil-like conformation.
Fig. 3.
Mean-squared atomic displacements obtained from the elastic neutron-scattering intensities from hydrated lysozyme as a function of temperature
Although we do not suggest that the conformational flexibility that accompanies enhanced atomic dynamics is not important for biological activity [44–46], our data indicate that the onset of the transition in the mean-squared displacement alone is not sufficient for explaining the onset of biological function. It is widely believed that the transition to anharmonicity is driven by the hydration water, which “slaves” the motions of biomolecules. However, the exact mechanism of such “enslavement” has been extensively debated. For example, it has been reported that the transition is coupled to the onset of translational motions in hydration water [47–49]. In addition, it has been debated whether the transition is a real phenomenon, or merely a manifestation of the fact that the measurable atomic dynamics enter the experimentally accessible resolution window above certain temperatures [17, 18, 24, 25, 27]. In such a scenario, the dynamic motions of the biomolecules may increase without any discontinuity. Although the detailed mechanism responsible for the transition remains in question (see Doster [50] for a recent discussion), it should be noted that various hypotheses are not necessarily mutually exclusive. For instance, it has been suggested that hydration water, in general, exhibits a dynamical transition unless the hydration level is too low [51]. Furthermore, the component that gives rise to this transition is physically similar to the translational component in that it requires simultaneous breaking of several hydrogen bonds of a hydration water molecule. However, it has the appearance of a β component because of the localized nature of the motions; on a neutron backscattering spectrometer, this component can be resolved above approximately 200 K [51]. Thus, the first signs of the transition in hydrated biomolecules may coincide with the entrance of this component to the resolution window. At 220–230 K, the hydration water experiences a dynamical transition [20–23] that further increases anharmonicity. Finally, the onset of long-range translational motions of hydration water leads to an even faster increase in anharmonicity at temperatures above 240 K [47–49]. Thus, several different phenomena could contribute to the experimentally observed increase in the mean-squared displacements.
Recent studies of protein dynamics in solutions and solid environments [26, 28] have demonstrated that the internal protein motions are slaved to the β fluctuations of the hydration shell, whereas the large-scale protein motions are slaved to the fluctuations of the bulk solvent. In the framework of this idea, the transition in the mean-squared atomic displacements can be expected for any protein, whether native or denatured, as long as the protein is sufficiently hydrated. Thus, the results of the present study are consistent with the dominant role in the transition of the mean-squared displacements played by the fluctuations of the hydration shell.
Acknowledgements
The authors are thankful to K. W. Herwig and C. Hoffmann for critical reading of the manuscript, and K. L. Weiss for useful technical discussions. This work was supported by the US Department of Energy Basic Energy Sciences and the Office of Biological and Environmental Research, using facilities supported by Oak Ridge National Laboratory, and managed by UT-Battelle, LLC, for the US DOE under Contract No. DE-AC05-00OR22725.
References
- 1.Parak F, Formanek H.Study on vibration and crystal lattice fault of temperature factor in myoglobin by comparative Mossbauer absorption measurements with X-ray structure data Acta Crystallogr. A 197127573–578. 10.1107/S05677394710012811971AcCrA..27..573P [DOI] [Google Scholar]
- 2.Keller H, Debrunner PG.Evidence for conformational and diffusional mean-square displacements in frozen aqueous-solution of oxymyoglobin Phys. Rev. Lett. 19804568–71. 10.1103/PhysRevLett.45.681980PhRvL..45...68K [DOI] [Google Scholar]
- 3.Parak F, Knapp EW, Kucheida D. Protein dynamics–Mossbauer–spectroscopy on deoxymyoglobin crystals. J. Mol. Biol. 1982;161:177–194. doi: 10.1016/0022-2836(82)90285-6. [DOI] [PubMed] [Google Scholar]
- 4.Doster W, Cusack S, Petry W.Dynamical transition of myoglobin revealed by inelastic neutron-scattering Nature 1989337754–756. 10.1038/337754a01989Natur.337..754D [DOI] [PubMed] [Google Scholar]
- 5.Paciaroni A, Cinelli S, Cornicchi E, Francesco A, Onori G.Fast fluctuations in protein powders: the role of hydration Chem. Phys. Lett. 2005410400–403. 10.1016/j.cplett.2005.05.0982005CPL...410..400P [DOI] [Google Scholar]
- 6.Cornicchi E, Onori G, Paciaroni A.Picosecond-time-scale fluctuations of proteins in glassy matrices: the role of viscosity Phys. Rev. Lett. 200595158104. 10.1103/PhysRevLett.95.1581042005PhRvL..95o8104C [DOI] [PubMed] [Google Scholar]
- 7.Roh JH, Novikov VN, Gregory RB, Curtis JE, Chowdhuri Z, Sokolov AP.Onsets of anharmonicity in protein dynamics Phys. Rev. Lett. 200595038101. 10.1103/PhysRevLett.95.0381012005PhRvL..95c8101R [DOI] [PubMed] [Google Scholar]
- 8.Roh JH, Curtis JE, Azzam S, Novikov VN, Peral I, Chowdhuri Z, Gregory RB, Sokolov AP.Influence of hydration on the dynamics of lysozyme Biophys. J. 2006912573–2588. 10.1529/biophysj.106.0822142006BpJ....91.2573R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornicchi E, Marconi M, Onori G, Paciaroni A.Controlling the protein dynamical transition with sugar-based bioprotectant matrices: a neutron scattering study Biophys. J. 200691289–297. 10.1529/biophysj.106.0817522006BpJ....91..289C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caliskan G, Briber RM, Thirumalai D, Garcia-Sakai V, Woodson SA, Sokolov AP. Dynamic transition in tRNA is solvent induced. J. Am. Chem. Soc. 2006;128:32–33. doi: 10.1021/ja056444i. [DOI] [PubMed] [Google Scholar]
- 11.Cornicchi E, Capponi S, Marconi M, Onori G, Paciaroni A.Temperature dependence of fast fluctuations in single- and double-stranded DNA molecules: a neutron scattering investigation Philos. Mag. 200787509–515. 10.1080/147864306009090222007PMag...87..509C [DOI] [Google Scholar]
- 12.Roh JH, Briber RM, Damjanovic A, Thirumalai D, Woodson SA, Sokolov AP.Dynamics of tRNA at different levels of hydration Biophys. J. 2009962755–2762. 10.1016/j.bpj.2008.12.38952009BpJ....96.2755R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel RM, Smith JC, Ferrand M, Hery S, Dunn R, Finney JL. Enzyme activity below the dynamical transition at 220 K. Biophys. J. 1998;75:2504–2507. doi: 10.1016/S0006-3495(98)77694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel RM, Finney JL, Reat V, Dunn R, Ferrand M, Smith JC. Enzyme dynamics and activity: time-scale dependence of dynamical transitions in glutamate dehydrogenase solution. Biophys. J. 1999;77:2184–2190. doi: 10.1016/S0006-3495(99)77058-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolov AP, Grimm H, Kisliuk A, Dianoux AJ. Slow relaxation process in DNA. J. Biol. Phys. 2001;27:313–327. doi: 10.1023/A:1014228824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG.Slaving: Solvent fluctuations dominate protein dynamics and functions Proc. Natl. Acad. Sci. USA 20029916047–16051. 10.1073/pnas.2126378992002PNAS...9916047F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel RM, Finney JM, Smith JC. The dynamic transition in proteins may have a simple explanation. Faraday Discuss. 2002;122:163–169. doi: 10.1039/b200989g. [DOI] [PubMed] [Google Scholar]
- 18.Becker T, Hayward JA, Finney JL, Daniel RM, Smith JC.Neutron frequency windows and the protein dynamical transition Biophys. J. 2004871436–1444. 10.1529/biophysj.104.0422262004BpJ....87.1436B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenimore PW, Frauenfelder H, McMahon BH, Young RD.Bulk-solvent and hydration-shell fluctuations, similar to alpha- and beta-fluctuations in glasses, control protein motions and functions Proc. Natl. Acad. Sci. USA 200410114408–14413. 10.1073/pnas.04055731012004PNAS..10114408F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SH, Liu L, Fratini E, Baglioni P, Faraone A, Mamontov E.Observation of fragile-to-strong dynamic crossover in protein hydration water Proc. Natl. Acad. Sci. USA 20061039012–9016. 10.1073/pnas.06024741032006PNAS..103.9012C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Yan Z, Xu L, Mazza MG, Buldyrev SV, Chen SH, Sastry S, Stanley HE.Glass transition in biomolecules and the liquid-liquid critical point of water Phys. Rev. Lett. 200697177802. 10.1103/PhysRevLett.97.1778022006PhRvL..97q7802K [DOI] [PubMed] [Google Scholar]
- 22.Chen SH, Liu L, Chu X, Zhang Y, Fratini E, Baglioni P, Faraone A, Mamontov E.Experimental evidence of fragile-to-strong dynamic crossover in DNA hydration water J. Chem. Phys. 2006125171103. 10.1063/1.23724912006JChPh.125q1103C [DOI] [PubMed] [Google Scholar]
- 23.Chu XQ, Fratini E, Baglioni P, Faraone A, Chen SH.Observation of a dynamic crossover in RNA hydration water which triggers a dynamic transition in the biopolymer Phys. Rev. E 200877011908. 10.1103/PhysRevE.77.0119082008PhRvE..77a1908C [DOI] [PubMed] [Google Scholar]
- 24.Khodadadi S, Pawlus S, Roh JH, Garcia-Sakai V, Mamontov E, Sokolov AP.The origin of the dynamic transition in proteins J. Chem. Phys. 2008128195106. 10.1063/1.29278712008JChPh.128s5106K [DOI] [PubMed] [Google Scholar]
- 25.Khodadadi S, Pawlus S, Sokolov AP. Influence of hydration on protein dynamics: combining dielectric and neutron scattering spectroscopy data. J. Phys. Chem. B. 2008;112:14273–14280. doi: 10.1021/jp8059807. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Fenimore PW, Frauenfelder H, Mezei F, Swenson J, Young RD. Protein fluctuations explored by inelastic neutron scattering and dielectric relaxation spectroscopy. Philos. Mag. 2008;88:3877–3883. doi: 10.1080/14786430802585117. [DOI] [Google Scholar]
- 27.Sokolov AP, Roh JH, Mamontov E, Garcia-Sakai V.Role of hydration water in dynamics of biological macromolecules Chem. Phys. 2008345212–218. 10.1016/j.chemphys.2007.07.0132008CP....345..212S [DOI] [Google Scholar]
- 28.Frauenfelder H, Ghen G, Berendzen J, Fenimore PW, Jansson H, McMahon BH, Stroe IR, Swenson J, Young RD.A unified model of protein dynamics Proc. Natl. Acad. Sci. USA 20091065129–5134. 10.1073/pnas.09003361062009PNAS..106.5129F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parak F, Knapp EW.A consistent picture of protein dynamics Proc. Natl. Acad. Sci. USA 1984817088–7092. 10.1073/pnas.81.22.70881984PNAS...81.7088P [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen BF, Stock AM, Ringe D, Petsko GA.Crystalline ribonuclease-A loses function below the dynamic transition at 220K Nature 1992357423–424. 10.1038/357423a01992Natur.357..423R [DOI] [PubMed] [Google Scholar]
- 31.Ferrand M, Dianoux AJ, Petry W, Zaccai G.Thermal motions and function of bacteriorhodopsin in purple membranes—effects of temperature and hydration studied by neutron-scattering Proc. Natl. Acad. Sci. USA 1993909668–9672. 10.1073/pnas.90.20.96681993PNAS...90.9668F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostermann A, Waschipky R, Parak FG, Nienhaus GU.Ligand binding and conformational motions in myoglobin Nature 2000404205–208. 10.1038/350046222000Natur.404..205O [DOI] [PubMed] [Google Scholar]
- 33.Dunn RV, Reat V, Finney J, Ferrand M, Smith JC, Daniel RM. Enzyme activity and dynamics: xylanase activity in the absence of fast anharmonic dynamics. Biochem. J. 2000;346:355–358. doi: 10.1042/0264-6021:3460355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bragger JM, Dunn RV, Daniel RM. Enzyme activity down to −100 degrees C. Biochim. Biophys. Acta. 2000;1480:278–282. doi: 10.1016/s0167-4838(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 35.He YF, Ku PI, Knab JR, Chen JY, Markelz AG.Protein dynamical transition does not require protein structure Phys. Rev. Lett. 2008101178103. 10.1103/PhysRevLett.101.1781032008PhRvL.101q8103H [DOI] [PubMed] [Google Scholar]
- 36.Mamontov E, Zamponi M, Hammons S, Keener WS, Hagen M, Herwig KW, BASIS A new backscattering spectrometer at the SNS. Neutron News. 2008;19:22–24. doi: 10.1080/10448630802210578. [DOI] [Google Scholar]
- 37.Mamontov E, Luo H, Dai S. Proton dynamics in N,N,Nʹ,Nʹ-Tetramethylguanidinium Bis(perfluoroethylsulfonyl)imide protic ionic liquid probed by quasielastic neutron scattering. J. Phys. Chem. B. 2009;113:159–169. doi: 10.1021/jp808102k. [DOI] [PubMed] [Google Scholar]
- 38.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochem. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 39.Stokkum IHM, Spoelder HJW, Bloemendal M, Grondelle R, Groen FCA. Estimation of protein secondary structure and error analysis from CD spectra. Anal. Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-Q. [DOI] [PubMed] [Google Scholar]
- 40.Whitmore L, Wallace BA. Protein secondary structure analysis from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 41.Bée M. Quasielastic Neutron Scattering. Philadelphia: Adam Hilger; 1988. [Google Scholar]
- 42.Nakagawa H, Joti Y, Kitao A, Kataoka M.Hydration affects both harmonic and anharmonic nature of protein dynamics Biophys. J. 2008952916–2923. 10.1529/biophysj.107.1285462008BpJ....95.2916N [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Receveur V, Calmettes P, Smith JC, Desmadril M, Coddens G, Durand D. Picosecond dynamical changes on denaturation of yeast phosphoglycerate kinase revealed by quasielastic neutron scattering. Proteins: Struct. Funct. Bioinformatics. 1997;28:380–387. doi: 10.1002/(SICI)1097-0134(199707)28:3<380::AID-PROT8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 44.Zaccai G.How soft is a protein? A protein dynamics force constant measured by neutron scattering Science 20002881604–1607. 10.1126/science.288.5471.16042000Sci...288.1604Z [DOI] [PubMed] [Google Scholar]
- 45.Parak FG.Physical aspects of protein dynamics Rep. Prog. Phys. 200366103–129. 10.1088/0034-4885/66/2/2012003RPPh...66..103P [DOI] [Google Scholar]
- 46.Frauenfelder H, Fenimore PW, Chen G, McMahon BH.Protein folding is slaved to solvent motions Proc. Natl. Acad. Sci. USA 200610315469–15472. 10.1073/pnas.06071681032006PNAS..10315469F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai AM, Neumann DA, Bell LN. Molecular dynamics of solid-state lysozyme as affected by glycerol and water: a neutron scattering study. Biophys. J. 2000;79:2728–2732. doi: 10.1016/S0006-3495(00)76511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarek M, Tobias DJ.Role of protein-water hydrogen bond dynamics in the protein dynamical transition Phys. Rev. Lett. 200288138101. 10.1103/PhysRevLett.88.1381012002PhRvL..88m8101T [DOI] [PubMed] [Google Scholar]
- 49.Tournier AL, Xu J, Smith JC. Translational hydration water dynamics drives the protein glass transition. Biophys. J. 2003;85:1871–1875. doi: 10.1016/S0006-3495(03)74614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doster W. The dynamical transition of proteins, concepts and misconceptions. Eur. Biophys. J. 2008;37:591–602. doi: 10.1007/s00249-008-0274-3. [DOI] [PubMed] [Google Scholar]
- 51.Mamontov E, Vlcek L, Wesolowski DJ, Cummings PT, Rosenqvist J, Wang W, Cole DR, Anovitz LM, Gasparovic G.Suppression of the dynamic transition in surface water at low hydration levels: a study of water on rutile Phys. Rev. E 200979051504. 10.1103/PhysRevE.79.0515042009PhRvE..79e1504M [DOI] [PubMed] [Google Scholar]