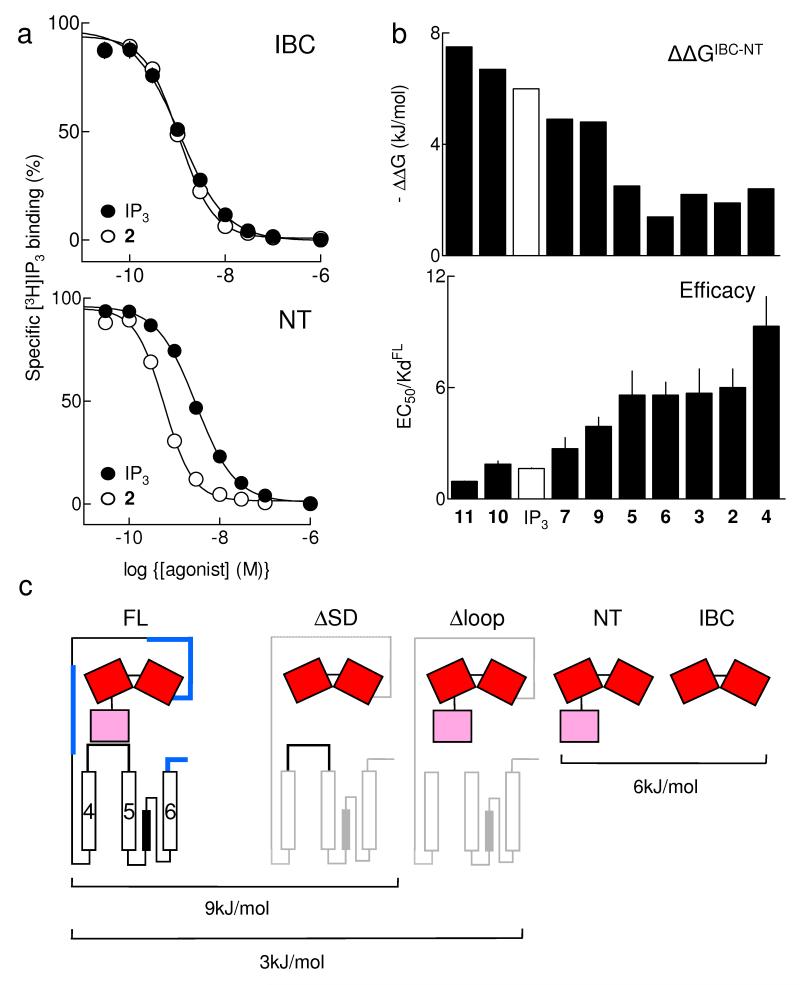
Figure 3.
Partial agonists are IP3-like in their interactions with the IBC. (a) Equilibrium-competition binding to IBC (top) and NT (bottom) with 3H-IP3 and either IP3 or 2, n ≥ 17. (b) ΔΔG (ΔGIBC-ΔGNT), reflecting ΔG used to rearrange the IBC-SD relationship, is shown for each ligand, and compared with the efficacy of each (EC50/Kd, with Kd determined for full-length IP3R1). Results (a,b) are means ± SEM. (c) Estimated ΔG for conformational changes associated with IP3R activation. The affinity (Kd) of IP3 for IP3R1 truncated as shown was measured herein (Table 1) or by others: by4 for ΔSD (IP3R1 lacking residues 1-223), and by32 for Δloop (IP3R1 lacking residues 2428-2437); ΔG was then calculated from ΔG = -RTlnKd. The Kd for IP3 was not directly measured in32, but under the conditions used the 4-fold increase in IP3 binding after deletion of residues 2428-2437 (ie Δloop) is likely to reflect a 4-fold decrease in Kd. We assume that deletion of IP3R fragments through which conformational changes must pass increases IP3 affinity because less binding energy is diverted into re-arranging the protein (Fig. 1b). Deletions of many other regions (shown in blue) do not increase IP3 affinity4, suggesting that the IP3-evoked conformational changes do not pass through them. This analysis is consistent with each IP3 binding event diverting ~9kJ/mol into conformational changes of the IP3R, of which ~6kJ/mol rearranges the SD-IBC relationship, and ~3kJ/mol is used by the SD to gate the pore.