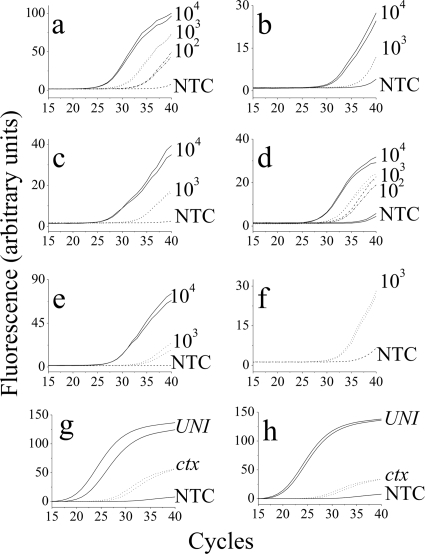
FIG. 5.
Real-time PCR amplification for the detection of V. cholerae O1. (a to c) V. cholerae O1 (102 to 104 cells) in 1 liter saline was run through a ROAK-packed column and eluted with 1.8 ml 70% ethanol. After DNA extraction, 25% of cells were amplified. Shown are relative SYBR green fluorescence developments as a function of cycle number. Samples were amplified using 16S universal primers (UNI) for detection of bacteria (a) and V. cholerae-specific primers, ompW (b) and ctx (c) locus primers. (d to f) V. cholerae O1 in tap water was analyzed as described above. (d and e) Analysis of 1 liter tap water when amplified with ompW and with ctx, respectively. (f) Results obtained with 10 liters tap water inoculated with 103 V. cholerae O1 cells (10 CFU/100 ml) and amplified with ctx locus primers. NTC is a nontemplate control. (g and h) V. cholerae O1 detection against a high background of bacteria. Analysis of 1 liter tap water inoculated with 100 CFU/100 ml of V. cholerae and of a mix of 104 CFU/100 ml defined bacteria (E. faecalis, S. aureus, and E. coli) (g) and undefined bacteria (sewage water after treatment) (h), when amplified with UNI and with ctx locus primers. In all panels, an absence of the second (duplicate) line indicates consistency.