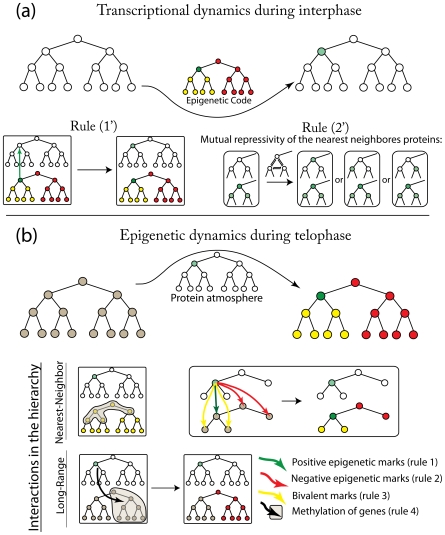
Figure 3. Rules that govern interactions within epigenetic and genetic networks.
(a) During interphase, gene expression profiles of master-regulatory modules are established. Gene expression is influenced by epigenetic marking of the corresponding gene and interactions between expressed proteins. Two rules reflect this in our simulation: 1) when master-regulatory gene is in epigenetically marked positively, it favors expression of the corresponding protein; 2) when two (three) neighboring genes are in epigenetically open states, they all favor expression of corresponding proteins, but due to their mutually repressive action (see text) only one of two(three) genes are expressed. Which gene is expressed is chosen stochastically. (b) During the telophase, the protein environment can alter the epigenetic marks on the master-regulatory genes. Epigenetic marks on both neighboring and distant genes in the hierarchy can be altered. Long-range effect is typically mediated through DNA methylation which epigenetically silences all of the master-regulatory genes of unrelated lineages and also ancestral states (see text). Short-range interactions affect nearest-neighbors differentially: progenies master-regulatory genes are preferentially put into bivalent states while progenitor and competing lineage modules are epigenetically silenced. The color code representing genetic and epigenetic states is the same as in Fig. 1. The numbers corresponding to the rules are the same as in text and Table 1.