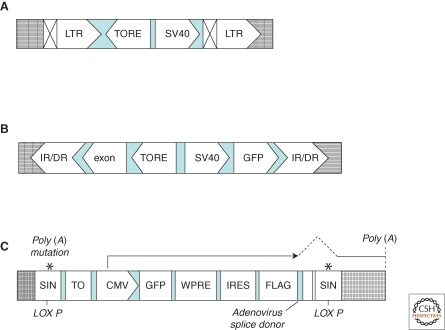
Figure 2.
Structures of different promoter insertion vectors. (A) Retroviral vectors. Retroviral vectors optimized for insertional mutagenesis were generated by using the MMLV (Moloney murine leukemia virus) backbone of pBabe vectors. The recognition sequence for Cre recombinase was placed inside the 3′ LTR so that, on reverse transcription, this motif would be copied into the 5′-LTR (Kandel et al. 2005). Abbreviations: TORE, tetracycline-regulated promoter; SV40, promoter and enhancer from the SV40 virus; LTR, long terminal repeat. Symbols: ⊠, the LTR modification, including a LoxP site for cre-mediated recombination, leading to deletion of the promoter; grid lines, host genomic DNA. (B) Sleeping Beauty transposon-based vectors. This series of mutagenesis vectors contain a coding region for GFP controlled by TORE, which is placed in the vicinity of a divergent SV40 promoter, followed by a mini-exon, which ends with an unpaired adenoviral splice site (Kandel et al. 2005). These vectors can be cotransfected with the Sleeping Beauty transposase to randomly generate mutations. Because the promoter is tet-regulated, mutant phenotypes caused by the inserted promoter can be reverted on promoter shutdown on treatment with the tetracycline analog doxycycline (Dasgupta et al. 2008). Abbreviations: IR/DR, terminal repeat from Sleeping Beauty; TORE, modified tetracycline-regulated promoter; GFP, coding region of enhanced GFP. Symbol: grid lines, host genomic DNA. (C) VBIM vectors. The VBIM vectors use a lentiviral backbone with polyadenylation mutations in both 5′ and 3′ LTRs and a lox P site in the 3′ LTR. This design allows excision of all but 238 bp of inert proviral DNA, lacking both promoter activity and polyadenylation signals, following cleavage by Cre, a critical feature for complete and consistent phenotypic reversion. The polyadenylation mutations also permit the mutagenic promoter to be placed in the same direction as transcription from the 5′ LTR during virus packaging, resulting in high virus titers that are comparable to those obtained with standard lentiviral vectors, eliminating promoter conflicts that occur with alternative designs. Furthermore, eliminating promoter interference also permits the use of a strong full-length CMV promoter rather than a minimal tetracycline-regulated promoter, which requires the tet activator protein tTA to be present in the target cells (Kandel et al. 2005). Thus, primary and even differentiated or senescent cells can be mutated without prior manipulation to express tTA or an ecotropic receptor, as previously required (Kandel et al. 2005). Besides these, a tetracycline-binding element upstream of the full-length CMV promoter is introduced to allow tetracycline-regulated control of the mutagenic promoter after the mutant has been created, by using a TR-KRAB fusion protein. Detailed description of this series of vectors has been reported recently (Lu et al. 2009a). Abbreviations: CMV, cytomegalovirus promoter; GFP, green fluorescent protein; IRES, internal ribosome entry sequence; LoxP, site for cre-mediated recombination; SIN, self-inactivating LTR; TO, tetracycline operon; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. Symbol: grid lines, host genomic DNA.