Abstract
A common ancestor to the three p53 family members of human genes p53, p63, and p73 is first detected in the evolution of modern‐day sea anemones, in which both structurally and functionally it acts to protect the germ line from genomic instabilities in response to stresses. This p63/p73 common ancestor gene is found in almost all invertebrates and first duplicates to produce a p53 gene and a p63/p73 ancestor in cartilaginous fish. Bony fish contain all three genes, p53, p63, and p73, and the functions of these three transcription factors diversify in the higher vertebrates. Thus, this gene family has preserved its structural features and functional activities for over one billion years of evolution.
The p53 tumor suppressor probably arose from an ancestral gene that protected the germ line against genomic instability and duplicated to give p53 and p63/p73 in cartilaginous fish.
In this article, the structure and the function of the p53 family of genes are reviewed over one billion years of evolution. The origins of a p53 ancestor gene are first clearly observed in the modern-day descendants of both the single cell choanoflagellates and the early metazoan sea anemone. In these organisms, the ancestral gene is most closely related to a combined p63/p73-like gene. Remarkably, the function of this ancestral gene in the sea anemone is to protect the germ-line gametes from DNA damage. This function persists in the insects, worms, clams, vertebrates, and humans; thus, this important function is over one billion years old. In the early vertebrate lineage, characterized by cartilaginous fish, this ancestral gene is duplicated and a gene most closely related to the p53 gene is produced. This is a time when somatic stem cells are employed to regenerate adult tissues and the p53 gene takes on the related function of a tumor suppressor, protecting these somatic stem cells and progenitor cells from mutations that arise from DNA damage. Thus, the p53 gene acquires functions in somatic cells that the ancestor gene performs in the germ line.
With the development of bony fish, there is a second duplication of the ancestor gene, producing a p63 and a p73 gene. As vertebrates develop, there is evidence that the functions of these genes enlarge and change into transcriptional regulators for the production of skin and other organs (p63), and have a role in the formation of parts of the immune and nervous systems (p73). From bony fish through amphibians, reptiles, and mammals, there is a dramatic increase in the size of the introns of the p63 gene and, to a lesser extent, the p73 gene, whereas the p53 gene remains quite small. The separate p63 and p73 genes never quite lose their original functions. Experiments with knockout mice indicate that mammals continue to employ p63 and p73 for the surveillance of the genomic integrity of the female germ line. Here, again, there are striking similarities between the stress signals that activate the p63/p73 (as well as p53) genes in the germ line of worms and mammals, resulting in both cases in apoptosis. There is some evidence that the p63 ancestor gene was under strong selective pressure at the time the most advanced reptiles (giving birth to live young rather than laying eggs) began to develop into mammals. There is evidence that selective pressures upon alleles in the p73 gene were acting as mammals diversified. Strong selection pressures are observed for selected alleles in the p53 gene in the last 30,000–50,000 years of human evolution, with the appearance of Asians and Caucasians in the human population pools.
This reconstruction of the evolutionary history of the three sister genes that form the p53 family of transcription factors shows the importance of germ line and stem cell reproductive fidelity during one billion years of evolution. The structural and functional conservation of this property helps to explain why these proteins play such a central role in biology. This history also elucidates how gene duplication and diversification can occur and how the new products fit into regulatory circuits closely related to those observed in more primitive organisms. Thus, the AKT and Abl kinases play an important role in the regulation of this p53/p63 and p73 family of gene products in both worms and humans. Clearly, evolutionary forces have shaped the p53, p63, and p73 genes. In turn, the functions of these genes shaped the fidelity of the offspring produced and guided the variations permitted so as to allow natural selection to act upon these progeny.
THE STRUCTURE OF THE p53, p63, AND p73 GENES IN HUMANS
The human p53 gene is located at chromosome 17p13.1. It is composed of 19,198 nucleotides from the first exon to the 11th exon. The coding sequence begins in the second exon and ends in the last exon (Table 1). The p63 gene is located on chromosome 3q27-29. It is composed of 265,822 nucleotides divided up into 14 exons. The coding sequence can start in exon one and continue through exon 14. (Table 1). The p73 gene is located on chromosome 1p36.3. It is composed of 80,728 nucleotides divided up into 14 exons. The coding sequence can start in exon 2 and end in exon 14 (Table 1). None of these dimensions take into account the regulatory regions that control the synthesis of these gene products. The larger sizes of the p63 and p73 genes derive from much larger introns and a few extra exons. This dramatic increase in intron size in vertebrates is first observed after the split from the bony fishes. This has permitted higher levels of recombination to occur in the p63 gene and intermediate levels in the p73 gene compared with the recombination levels in the p53 gene in these higher vertebrates. This in turn gives rise to an increased number of haplotypes in the p63 gene and p73 gene and greater diversity of haplotypes (Table 2).
Table 1.
Exon structure of human p53-family genes.
| p53 | p63 | p73 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Full gene length | 19,198 | 265,822 | 80,728 | ||||||
| Coding region | Starts in exon two, goes through exon 11 | Starts in exon one, goes through exon 14 | Starts in exon two, goes through exon 14 | ||||||
| Exons | start | end | length | start | end | length | start | end | length |
| Exon 1 | 1 | 223 | 223 | 1 | 151 | 151 | 1 | 77 | 77 |
| Exon 2 | 10,978 | 11,079 | 102 | 106,314 | 106,442 | 129 | 29,769 | 29,866 | 98 |
| Exon 3 | 11,197 | 11,218 | 22 | 107,216 | 107,348 | 133 | 30,496 | 30,616 | 121 |
| Exon 4 | 11,328 | 11,606 | 279 | 176,846 | 177,100 | 255 | 54,985 | 55,227 | 243 |
| Exon 5 | 12,364 | 12,547 | 184 | 232,806 | 232,992 | 187 | 69,457 | 69,643 | 187 |
| Exon 6 | 12,629 | 12,741 | 113 | 235,256 | 235,371 | 116 | 70,790 | 70,905 | 116 |
| Exon 7 | 13,310 | 13,419 | 110 | 236,407 | 236,516 | 110 | 74,551 | 74,660 | 110 |
| Exon 8 | 13,763 | 13,899 | 137 | 237,154 | 237,290 | 137 | 75,064 | 75,206 | 143 |
| Exon 9 | 13,992 | 14,065 | 74 | 237,898 | 237,980 | 83 | 75,565 | 75,653 | 89 |
| Exon 10 | 16,885 | 16,991 | 107 | 241,433 | 241,569 | 137 | 76,763 | 76,884 | 122 |
| Exon 11 | 17,910 | 19,198 | 1289 | 254,968 | 255,125 | 158 | 77,436 | 77,584 | 149 |
| Exon 12 | 257,914 | 258,058 | 145 | 78,363 | 78,501 | 139 | |||
| Exon 13 | 259,363 | 259,456 | 94 | 78,899 | 78,992 | 94 | |||
| Exon 14 | 262,780 | 265,822 | 3043 | 80,183 | 80,728 | 546 | |||
Exons and coding region for the longest transcript were extracted from Ensembl database, release 54 (Hubbard et al. 2009).
Table 2.
Summary of gene diversity for p53 family in human populations from HapMap Phase 3.
| Gene symbol | Populationa | Sample size (N)b | SNP number | Haplotype number | Haplotype diversityc |
|---|---|---|---|---|---|
| TP53 | African | 399 | 9 | 22 | 0.879 |
| Caucasian | 201 | 7 | 10 | 0.520 | |
| Asian | 343 | 10 | 19 | 0.716 | |
| TP63 | African | 399 | 167 | 696 | 0.999 |
| Caucasian | 201 | 161 | 386 | 1.000 | |
| Asian | 343 | 159 | 664 | 1.000 | |
| TP73 | African | 399 | 47 | 496 | 0.998 |
| Caucasian | 201 | 44 | 251 | 0.994 | |
| Asian | 343 | 46 | 506 | 0.998 |
a Three major populations of different ancestry in HapMap Phase 3: Africans include ASW (African ancestry in Southwest USA), LWK (Luhya in Webuye, Kenya), MKK (Maasai in Kinyawa, Kenya), and YRI (Yoruba in Ibadan, Nigeria); Caucasians include CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) and TSI (Toscans in Italy); Asians include CHB (Han Chinese in Beijing, China), CHD (Chinese in Metropolitan Denver, Colorado), GIH (Gujarati Indians in Houston, Texas) and JPT (Japanese in Tokyo, Japan).
b Haplotypes with missing data were not included.
c Haplotype diversity, the expected heterozygosity based on haplotype frequencies, was estimated using DnaSP (v5) software.
Based on the HapMap-3 results, the p53 gene in Africans has 22 haplotypes and a haplotype diversity index of 0.879. In Caucasians, there are 10 haplotypes with a diversity of 0.520, and in Asians there are 19 haplotypes with a diversity index of 0.716. This shows a dramatic selection (lowering diversity) of a haplotype in Caucasians and to a lesser extent in Asians (it is the codon 72 arginine haplotype that is being selected for in these populations) with a considerable lowering of diversity (Table 2). In contrast, the p63 gene of Africans has 696 haplotypes and a diversity index of 0.999. Caucasians have 386 haplotypes and a diversity index of 1.000, and Asians have 664 haplotypes and a diversity index of 1.000 (very little linkage disequilibrium over the size of the entire gene but there is linkage disequilibrium between several SNPs within the gene). The p73 gene of Africans has 496 haplotypes with a haplotype diversity index of 0.998. Caucasians have 251 haplotypes with a diversity index of 0.994, and Asians have 506 haplotypes and a diversity index of 0.998 (Table 2). For the p53 gene, these numbers will depend upon the number of single‐nucleotide polymorphisms (SNPs) tested and the sample size (7–10 SNPs in 201–399 individuals times two chromosomes tested in each person). Even doubling the number of SNPs in the p53 genes tested (12–14 SNPs in 96 individuals or 192 chromosomes) gives the same result in the diversity index of Africans (0.943) and Caucasians (0.542) (H. Wang and A. Levine, unpubl. results).
Clearly, the diversity of the p53 gene in Caucasians is being restricted, probably because of the selection of the arginine allele of the codon 72 SNP. This allele (arginine compared with proline) has been shown to synthesize twofold more of the cytokine LIF, which is essential for the implantation of fertilized eggs into the uterus of mice and humans (Hu et al. 2007b; Hu et al. 2008; Shi et al, 2009; Kang et al. 2009). The minor allele of that SNP in Caucasians, the proline allele, has been shown to be over‐represented in women who have gone to an in vitro fertilization clinic because they have difficulties with the implantation of embryos into their uterus (Kang et al. 2009). Given that the p53 gene haplotype diversity index in Africans is high and the proline allele is not selected against, as in Caucasians and in Asians, the genetic background or additional alleles in the p53 pathway are important in this selection process. The specific alleles (codon 72) and haplotypes that are under selection in Caucasians has occurred recently, perhaps in the past 30,000 years, and is not relevant to evolutionary considerations over longer time frames.
As will become clear in this article, there has been a great deal of diversity and selective change in p53, p63, and p73 genes throughout a billion years of evolution, and especially in the vertebrates. A comparison of the rates of nucleotide or codon and amino acid changes has been employed to examine the evolution of the p63 and p73 genes in vertebrates (Pintus et al. 2007). Evidence for positive selection of p63 alleles was detected at the time of divergence of Synapsida and Reptiles. The Synapsida are mammal‐like reptiles or proto-mammals that are Amniotes and gave rise to live offspring (viviparity). Thus, the first vertebrates to develop their embryos within the females (not egg layers) were selecting for positive alleles in the p63 gene. Positive selection of p73 alleles occurred during diversification of mammals between the Carnavora (cats, bears, and dogs) and the Artiodactyla (pigs, sheep, and goats). Thus, indications of positive selection of alleles in the the p53, p63, and p73 genes have been found in the late stages of vertebrate evolution.
THE DOMAIN STRUCTURE OF THE p53, p63, AND p73 PROTEINS
Although this topic has been reviewed in detail in another article, it is useful to outline the domain structure of the p53 family proteins here because it bears upon the methods employed to compare these genes over large evolutionary time frames and points out the limitations of such studies. The human p53 gene encodes 393 amino acids for the full length isoform of the protein. The amino-terminal residues one to 42 contain a transactivation domain with a couple of residues (22 and 23) critical for this activity (Lin et al. 1994; Chang et al. 1995; Teufel et al. 2007). The negative regulator of p53, MDM-2, binds to these residues and induces an α-helical conformation, blocking transcription and ubiquitinating lysine residues in the carboxy-terminal domain region (Lin et al, 1994; Kussie et al. 1996; Rippin et al. 2002). A second transactivating domain at residues 43–63 presumably activates transcription of different p53‐regulated genes in the genome. This is followed by a proline‐rich domain (residues 61–94) that is involved in apoptosis and possibly protein–protein interactions that are mediated by SH-3 signals, PXXP (Walker and Levine 1996). Residues 102–292 make up the DNA binding domain that recognizes a degenerate DNA sequence near or in the introns of genes regulated by p53 (el-Deiry et al. 1992). The p53 protein functions as a dimer of a dimer and the tetramerization domain spans amino acid residues 324–355 (Jeffrey et al. 1995; Kitayner et al. 2006; Veprintsev et al. 2006). This is followed by a carboxy-terminal domain composed of residues 356–393, which regulates the stability and DNA binding activity of the p53 protein (Jayaraman and Prives 1995). Splicing at the amino-terminal end of the genes for p53, p63, and p73 gives rise to different isoforms that can act as repressors of transcription (deletion of the amino-terminal transactivation domains). Splicing at the carboxy-terminal ends of the p53, p63, and p73 genes give rise to isoforms that regulate stability of the protein and DNA binding (Pietsch et al. 2008; Yang et al. 1998; Melino et al. 2003; Moll and Slade 2004; Bourdon et al. 2005). Thus p53, p63, and p73 each have many different isoforms with a number of diverse properties. The domain organizations of p53, p63, and p73 are similar but the extended and different carboxy-terminal ends of p63 and p73 provide a larger diversity and more complex functions to those proteins (reviewed in a later section). Each of these three classes of proteins can regulate the transcription of a subset of genes in common and a subset of genes unique to each protein and presumably each isoform of each protein affects this specificity. The p53/p63 and p73 proteins bind to similar or identical DNA sequences, but there may well be quantitative differences between these family members and proteins in cells that modify their specificity to give rise to different regulatory functions known to exist (Ortt and Sinha 2006; Vigano et al. 2006; Smeenk et al. 2008; Tozluoglu et al. 2008).
Once the three genes, p53, p63, and p73, had been incorporated into the genomes of bony fish, the DNA binding domains of each of these genes evolved at quite different rates, indicating quite diverse functions (Table 3). For example, the rodents had a common ancestor with primates about 100 million years ago and the number of amino acid changes between human and rodent p53 protein in the DNA binding domain is 11–23, whereas for p63, it is zero and, for p73, it is three to four. By the elephant and the hyrax (the most distant mammals from humans), these numbers are p53/p63/p73; 19–28, zero, and six to seven. In bony fish, these numbers become p53 = 57–80 changes in amino acids, p63 = 7–11 changes, p73 = 18–21 changes. There were clearly different selection pressures on these three genes with p53 evolving at the fastest rate, p73 next, and p63 quite slowly. In the section of this article about the functional differences of these proteins, it will become clear that both the p63 and p73 functions took on important roles in development of the organism, whereas p53 was mainly employed for the surveillance of genome integrity of somatic stem cells and progenitor cells and plays less of an essential role in development of the embryo’s differentiated tissues.
Table 3.
Variation in the DNA binding domain of the p53 family genes in bony vertebrates.
| p53 | p63 | p73 | |
|---|---|---|---|
| Human | 0 | 0 | 0 |
| Chimp | 0 | 0 | |
| Gorilla | 1 | ||
| Orangutan | 1 | 0 | 0 |
| Macaque | 5 | 0 | 2 |
| Bushbaby | 4 | ||
| Gray Mouse Lemur | 11 | 0 | |
| Rat | 18 | 0 | 3 |
| Mouse | 23 | 0 | 4 |
| Kangaroo Rat | 1 | ||
| Squirrel | 0 | ||
| Guinea pig | 21 | 0 | 4 |
| Pika | 18 | ||
| Rabbit | 10 | 0 | |
| Dolphin | 15 | 0 | 6 |
| Cow | 18 | 4 | |
| Cat | 22 | ||
| Dog | 19 | 0 | 6 |
| Pteropus | 1 | 9 | |
| Horse | 0 | ||
| Hedgehog | 30 | 4 | |
| Elephant | 19 | 7 | |
| Hyrax | 28 | 0 | 6 |
| Opossum | 0 | ||
| Platypus | 0 | ||
| Zebra Finch | 0 | 7 | |
| Chicken | 0 | 6 | |
| Frog | 64 | 13 | |
| Stickleback | 80 | 11 | 21 |
| Medaka | 77 | 8 | 21 |
| Fugu | 76 | 10 | 21 |
| Tetraodon | 79 | 11 | 21 |
| Zebrafish | 57 | 7 | 18 |
Each value corresponds to the net amino acid difference between given species and human DNA binding domains (total of 195 amino acids). Only domains with complete sequences are shown. See Figure 1 for complete information on p53 family gene homologs.
THE LIMITATIONS OF EVOLUTIONARY STUDIES
To determine if an organism contains one, two, or three p53 family member genes, we rely upon protein sequences derived from the DNA sequences obtained from whole genome sequencing of the organism (Karolchik et al. 2008; Hubbard et al. 2009). However, some genomes have only a low DNA sequence coverage and are surely missing some percentage of genes, and others may have a poor assembly of the genome. Other genomes in the evolutionary tree have quite complete genome coverage and give accurate numbers of paralogs or homologs in a genome. One example of this is the p53 ortholog in chicken that has been identified and sequenced (Soussi et al. 1988) yet cannot be mapped to the current genome assembly despite high sequencing coverage. Thus, gene counts of p53 family members are estimates, except in those organisms with a very complete genome sequence. Second, this analysis compares related genes over a billion years of evolution. Over those distances in time, DNA sequences are quite degenerate. Only those amino acid residues that are highly selected for at identical or similar positions in a domain of a protein are truly conserved. This is true for the DNA binding domains of p53, p63, and p73, which are closely related to each other (paralogs) in an organism and closely related to their homologs between organisms. In the DNA binding domain, there are critical amino acids that contribute to the structure of the domain and make the DNA contacts that give specificity to the sequences recognized by this domain (based upon the crystal structure of both the Xenopus and the human p53 proteins) (Kussie et al. 1996; Wang et al. 1995). Indeed, the DNA binding domains from Drosophila and humans have very similar residues in identical positions and very similar predicted structures, and both will bind specifically to the same DNA sequence (Jin et al. 2000). The same is true for the p53‐like protein from Caenorhabditis elegans (Huyen et al. 2004). For this reason, the amino acid sequence of the DNA binding domains of p53, p63, and p73 were employed to search for the homologs of these genes in the genomes of the diverse organisms under study, with information about additional domains being used only when available (Lu and Abrams 2006; Ou et al. 2007). Although this is a conservative way to explore evolutionary relationships, it may detect p53‐related DNA binding domains that are functionally different from the human p53, p63, and p73. Moreover, all of the organisms that have been sequenced and are to be compared for the presence of p53 family genes donate DNA from only their contemporary representatives of each class of evolutionary organism. This means that some organisms may expand or contract the number of p53 family genes later in evolutionary times, well after the separation of common ancestors leading to more complex or higher organisms on a tree. Thus, the number of p53 family genes in an organism (1, 2, or 3) can arise from later duplications or deletions. For these reasons, the conclusions made about the evolutionary development of the p53 family of genes should not be considered a definitive statement, but rather a good estimate of past events.
THE PRESENCE OF HOMOLOGS OR PARALOGS OF p53 FAMILY GENES OVER EVOLUTIONARY TIME FRAMES
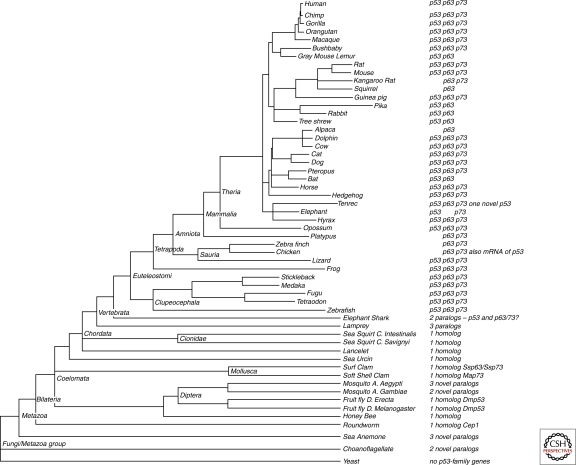
Figure 1 presents a consensus evolutionary tree spanning the early Metazoa through the primates (Hubbard et al. 2009; Sayers et al. 2009). What is clear from extensive DNA sequence data is that prokaryotes and yeast as a representative of early single-cell–multicellular (depending upon their environment and physiology) organisms have no detectible p53 family member genes. Representatives of the most primitive organisms with paralogs of p53/63/73 family member genes (it is not possible to know whether these genes are more closely related to p63 or p73, but the amino acid sequences and positions are not as close to p53) are the single-cell choanoflagellates (two paralogs) (Nedelcu and Tan 2007) and the sea anemone (three paralogs) (Pankow and Bamberger 2007). The sea anemone (and presumably related organisms the hydra and coral) Nematostella vectensis has a p63/p73 hybrid‐like gene (the precursor of p63 and p73) termed nvp-63 and it is present and functions in the germ‐line cells of these hydra (Pankow and Bamberger 2007). This is the earliest clear example of an ancestor family member in evolutionarily primitive organisms. Complete genome sequences from the sponges, which precede the sea anemones in evolution, are not yet available.
Figure 1.
Consensus phylogenetic tree and evidence of p53 family genes, reconstructed from Ensembl database, release 54, and NCBI taxonomy database. Tree branches are proportional to the genome-wide nucleotide substitution rates for jawed vertebrates (euteleostomi), but are qualitative for more primitive species. Evidence for p53 family genes was either extracted from Ensembl database or manually verified using BLAST (for the most recent assemblies available*) (Altschul et al. 1990; Hubbard et al. 2009; Sayers et al. 2009). Evidence for p53 family genes in clams comes from gene isolation by C. Walker and others (Kelley et al. 2001; Cox et al. 2003)
The first set of complete sequence information comes from the roundworms C. elegans and the insects like the honey bee and multiple species of the fruit fly Drosophila. All have a single homolog of the p53 family genes (Jin et al. 2000; Ollmann et al. 2000; Derry et al. 2001; Schumacher et al. 2001; Nedelcu and Tan 2007). This homolog is more closely related to a p63/p73 hybrid gene or ancestor gene and much less closely related to p53 itself (Ou et al. 2007). Some mosquitos have two or three paralogs of this p63/73 gene and show a tendency to duplicate it and possibly develop new functions (Nedelcu and Tan 2007). The sea urchin has a single homolog of this p63/p73 hybrid gene. Thanks to Charles Walker and his colleagues (Kelley et al. 2001; Jessen-Eller et al. 2002; Cox et al. 2003; Bottger et al. 2008; Holbrook et al. 2009; Siah et al. 2009), we know a great deal about the p53 gene in clams. Mya arenaria, a common clam species, has a single p63/p73 hybrid‐like gene but can splice the transcripts from this gene into a p53-like sequence and a p63/p73‐like sequence. The p53 isoform behaves like a functional p53 because clams can develop leukemia and in many cases p53 (in the p63/73 gene) mutations are found in the leukemic cells from this organism (Kelley et al. 2001; Bottger et al. 2008; Holbrook et al. 2009; Siah et al. 2009). It is also clear that a genera of clams, such as the common surf clam, has been exploring evolutionary expansions of splice variants of the p63/p73 gene, producing a p97 and a p120 set of isoforms with common DNA binding domains. Interestingly, the p120 isoform is expressed only in the embryonic stages of clam development and appears to be involved in neuronal functions (like p73 in mice) (Jessen-Eller et al. 2002; Cox et al. 2003).
As we move into the chordates and vertebrates, the Lancelet or Amphioxus (the earliest vertebrate) and the sea squirt have one clear homolog for the p63/p73 hybrid gene. Lampreys have three paralogs that are more difficult to classify. An example of a cartilaginous fish, the elephant shark, has two paralogs that are most like a p53 gene and a p63/p73 hybrid gene but this is an example of a difficult assembly and an incomplete genome sequence, so other family members of these genes remain possible. By the time we get to the bony fish (zebra, fugu, stickleback, etc.), clear homologs of p53, p63, and p73 genes are now observed (Cheng et al. 1997; Lee and Kimelman 2002; Pan et al. 2003). Thus, the bony fish are the first time in vertebrate evolution that the three gene family sisters p53, p63, and p73 are all clearly present together. This pattern of these three genes continues through the vertebrates right to present‐day primates, with some interesting exceptions. There is a group of closely related organisms, the rabbit, the tree shrew, and the pika, which appear to harbor only p53 and p63 homologs in their genomes and no p73 gene (Le Goas et al. 1997; Park et al. 2000; Hsueh et al. 2004). Similarly, the Lemur and the bat seem to have p53 and p63 homologs but no evidence for a p73 homolog. The elephant has a p53 homolog and a p73 homolog but no evidence for a p63 gene. There are even a few examples of a missing p53 homolog in the zebra finch, the platypus (p63 and p73), the kangaroo rat (p63 and p73), and the alpaca (p63 only). Whether these are examples of incomplete data for the analysis or a transfer of functions to other family members remains to be explored.
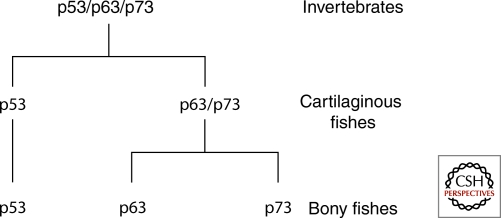
Casting aside these incomplete details, a reasonable pattern emerges from these data. Round worms and fruit flies appear to have a single gene that is a combination of p63 and p73. This gene appears as early as the sea anemone and possibly in the choanoflagellates. The clam has this same gene and employs it to produce different splice variants that resemble p53 and a p63/p73 hybrid gene. As evolution progresses up the ladder of complexity, organisms use this gene to expand new paralogs, possibly with different functions. The earliest Chordates also appear to have this single p63/p73 homolog. With the development of cartilaginous fish, there is a duplication of the p63/p73 gene, giving rise for the first time to a candidate p53 gene and a homolog of the p63/p73 hybrid gene (sometimes called p63 and sometimes called p73) (see Fig. 2). The evidence that a p53 gene separates from a p63/73 hybrid gene as a first step comes from its greater sequence separation from p63 and p73, which are much more closely related to each other (at the amino acid sequence level) than to p53 in these vertebrate genomes. This first separation of family members occurs in the early vertebrates (sea squirt and cartilaginous fish). A second separation of p63 from p73 is clearly observed in the bony fish and all subsequent higher organisms (Fig. 2). This genetic expansion of the family tree was surely followed by a differentiation of functions from which we can learn a great deal about modern‐day p53, p63, and p73 functions. The evolutionary requirements that first led to the creation and selection of a p63/p73 hybrid gene, followed by its duplication and the splitting off of the p53 gene, and finally the separation of p63 and p73 genes came about because the functions acquired by these genes over evolutionary time frames were useful. This is reviewed in the next section.
Figure 2.
Scenario of the p53 family gene evolution. The earliest indication of p53 family genes dates back to invertebrates and unicellular choanoflagellae. Aside from independent gene duplications, such as in sea anemone or mosquitos, the earliest indication for mammalian gene splitting dates back to cartilaginous fishes, with tentative orthologs visible in elephant sharks, and all three mammalian paralogs appearing in bony fishes.
THE EVOLUTION OF THE FUNCTIONS OF THE p53 FAMILY OF GENES
The most primitive organisms for which we have reliable functions for the p53 family genes are the starlet sea anemone (Nematostella vectensis), flies (Drosophila), and roundworms (Caenorhabditis). In the sea anemome, a p63/p73 hybrid gene (called nvp63) acts in the hydra gametes, responding to UV light and DNA damage by causing apoptosis (Pankow and Bamberger 2007). Thus, the most primitive organism with a p53 family gene member promotes apoptosis in response to DNA damage in the germ line. These hydra are among the earliest metazoans with a few differentiated cell types but have separated gamete production from the soma and they developed a p63/73 hybrid gene to watch over the fidelity of the germ line. In adult flies, the p63/p73 hybrid gene product (called dmp53) acts in the germ line to initiate apoptosis in response to DNA damage (Brodsky et al. 2000). The protein is a transcription factor that binds to DNA sequences closely related to those that the human p53, p63, and p73 proteins bind to in DNA. The Drosophila reaper and sickle genes are transcriptionally activated by this p63/p73 hybrid gene product and these gene products initiate apoptosis in germ cells with damaged DNA (Sogame et al. 2003).
During fly development, when cells are actively dividing, the absence of dmp53 can result in genomic instability, especially in response to DNA damage. Adult flies are largely postmitotic, with only a few cell types dividing (immune phagocytes, gut cells, and germ cells). Thus, the earliest functions for the p53 family genes are used to ensure germ‐line genome integrity and the larval stages of fly development, preventing birth defects and the production of abnormal embryos and flies. The fidelity of the developmental process ensures that resources are employed only for those faithfully reproducing the species. Similarly, in Caenorhabditis, the p63/73 hybrid gene (called CEP-1) acts during development and in the germ line of adults (Derry et al. 2001). In the germ line, CEP-1 responds with apoptosis to DNA damage, hypoxia, and starvation. It also acts, along with EGL-1 and CED-13 (a CEP-1 regulated gene), during oogenesis to decrease excess germ cell production by apoptosis to maintain homeostasis for egg production (Conradt and Horvitz 1998; Schumacher et al. 2005).
In the worm, CEP-1 is negatively regulated by the serine/threonine protein kinase AKT-1 (antiapoptotic) and the tyrosine kinase ABL-1 (there is no detectable MDM-2 in the genome) (Deng et al. 2004; Quevedo et al. 2007). Thus, the p63/73 gene in the worm has become integrated into pathways that sense glucose use, reproductive activities, oxygen availability, and genome integrity, and it responds by providing fidelity for developmental processes and germ-line production. The decreased expression of CEP-1 over the development and life of a worm results in an increased life span of these round worms (Arum and Johnson 2007; Tavernarakis et al. 2008). Here, we see for the first time the interplay between the availability of food in the environment, the homeostatic regulation of reproduction, the responses to a variety of stress signals that can alter the fidelity of the genome replication process, and the longevity of an organism. The first three (and possibly the fourth) of these functions are under intense evolutionary selection pressures. These selection pressures result from communication between p63/73 stress responses and the insulin-like growth factor (IGF-1) pathway, the communication of food resources with the control of egg production by p63/p73, the interactions between p63/73 and DNA repair pathways, and longevity regulated by the activity of the IGF-1 pathway (Nahor et al. 2005). This fixes in these organisms a temporal relationship between the age of attaining sexual maturity, the age at which reproductive activities decline because of an active homeostatic control (in the extreme menopause), and the longevity of a species. As cancers arise in the vertebrates, this, too, is controlled by these same genes and their signal transduction pathways.
As vertebrates emerged in evolution, they introduced a new body plan. Unlike flies and worms, which have adult organisms that are largely postmitotic with the exception of producing eggs and sperm for reproduction of the species, vertebrates developed somatic stem cells or progenitor cells that renew many tissues and organs of the body throughout life. Here, for the first time, the p63/73 hybrid gene is duplicated to produce a p53 gene. This new p53 gene now focuses upon the fidelity of somatic stem cell or progenitor cell duplication to prevent cancers arising from the high error rate that occurs during replication of damaged DNA. This is an extension of the germ‐line surveillance of the p63/73 hybrid gene observed in sea anemones, flies, and worms to somatic cells in fish. It is reasonable to assume that this new p53 gene and its tumor suppressor activity first appear in early vertebrates because some somatic cells (stem cells or progenitor cells) regenerate for the first time over the life of an organism. This new stem cell compartment is vulnerable to developing cancers because of environmental stresses. Thus, the expansion of the number of p53 family genes and selection for new functions arises from a gene dedicated to surveillance of germ‐line fidelity by a p63/73 hybrid ancestor into a p53 gene with the function of surveillance of DNA fidelity in somatic stem cells. This is a classical tumor-suppressor function.
With bony fish, amphibians, reptiles, mammals, and birds, the p63 and 73 genes separate (Fig. 2) and undergo positive selection so as to take on a set of functions critical to the differentiation of different tissues: p63 produces epithelial cell surfaces and selected organs, and p73 controls immunological and neuronal cells (Mills et al. 1999; Yang et al. 1999; Yang et al. 2000). There is good evidence that p63 and p73 gene functions continue in their role of female germ‐cell fidelity (Suh et al. 2006; Tomasini et al. 2008). Shortly after the separation of the p63 gene from the p73 gene, the insertions into the introns of the p63 gene altered its size dramatically. The size of the p63 gene in bony fish (zebrafish and the medaka) is about 25 to 35kb. In mammals it expanded in the platypus to 150kb, in the opossum to 234kb, and in humans to 265kb. This large increase in size results in a higher recombination frequency across the gene and a much greater number of haplotypes in a population (in Caucasians, the p53 gene has 10 haplotypes, whereas the p63 gene has 386 haplotypes). This creates a larger combination of haplotypes to be selected from in a population (see Table 2).
There are a large number of different knockout mice that have probed the functions of p63 and p73 genes in mice. It is clear that the p63 gene has evolved into a master regulator of epithelial cell production (Mills et al. 1999; Yang et al. 1999). P63 knockout mice are born without skin and related appendages (they die shortly after birth). They have craniofacial and limb abnormalities and have lost their prostate lineage cells (Mills et al. 1999; Yang et al. 1999; Signoretti et al. 2005). p63 is also employed in neuronal cell apoptosis (Jacobs et al. 2005). Isoform‐specific knockout mice show that p63 isoforms play a role in female infertility and even early menopause. p73 homozygous knockout mice can often die in utero and if born are runted and have a high mortality rate (Yang et al. 2000). They show a number of neurological defects including hippocampal dysgenesis and hydrocephalus. They have chronic inflammation and defects that lead to male infertility (lack of pherone detection). Specific isoform deletions of p73 show female infertility and less ovulated oocytes (Tomasini et al. 2008). Spindle abnormalities were common during oogenesis and genomic instability was observed. Thus, it is clear that in mammals, p63 and p73 acquire important developmental roles in programs that develop tissues.
At the same time, p63 and p73 retain their roles in germ‐line integrity and regulation of oogenesis. This is a function that p63/73 had in worms. These mouse studies make it equally clear that the heterozygous or homozygous loss of p63 or p73 show some tumor suppressor activity in selected tissues (Flores et al. 2005). Compound heterozygotes of p53/p63 or p53/p73 display a much higher tumor burden than p53 heterozygotes alone (Flores et al. 2005). In humans, p63 mutations (heterozygotes) result in ectrodactyly, ectodermal dysplasia, and cleft lip/palate or EEC syndrome (Celli et al. 1999; van Bokhoven et al. 2001). These mutations are also observed in families with nonsyndromic split‐hand/split‐foot malformations. These families do not have a Li-Fraumeni phenotype or a high cancer incidence, and somatic mutations in cancers contain few if any p63 or p73 mutations (Moll and Slade 2004).
THE ROLE OF p53, p63, AND p73 IN REGULATING DEVELOPMENT OF THE ORGANISM
There is abundant evidence that the p53, 63, and 73 family of genes regulates the male and female germ‐line production of gametes in response to stress, especially genomic damage. When male Drosophila (p63/73) or mice (p53) are irradiated and the DNA in cells undergoing spermatogenesis is damaged, there is a wave of apoptosis, killing these cells and causing a temporary sterility (Norimura et al. 1996; Ollmann et al. 2000). This is eventually replaced by sperm with nondamaged DNA that produce normal offspring. In this fashion, the p53 family of genes protects against developmental abnormalities in the progeny. Similarly, in worms (p63/p73) and mice (p63 and p73), the oocytes are regulated by these gene products so that DNA damage results in cell death (Suh et al. 2006; Tomasini et al. 2008). In addition to a stress signal composed of DNA damage, the sensing of food supplies in the environment is an important signal to regulate homeostasis of egg production, especially in organisms that do not have a lot of time to reproduce. The restriction of nutrition (glucose starvation) is sensed by the m-TOR pathway and can, under certain circumstances, activate the p53 pathway, leading to apoptosis and autophagy (Feng et al. 2005; Levine et al. 2006; Lee et al. 2007). In a reciprocal fashion, the activation of the p53 pathway induces the transcription of four gene products, PTEN, TSC-2, the β subunit of the AMP kinase gene, and the IGF-BP3 gene, which each modulate down the IGF-1 and mTor pathways, which in turn regulate growth and cell division through metabolic shifts (Feng et al. 2007).
The mTor kinase influences mitochondrial biogenesis, angiogenesis, ribosomal biogenesis, and cell size. The p53 pathway not only integrates these processes through modulating mTor activity, but it directly can modulate down aerobic glycolysis through the TIGAR gene product it regulates, and it modulates up oxidative phosphorylation through the control of the SCO2 gene (Bensaad et al. 2006; Matoba et al. 2006). The rapid cell growth during development requires a metabolic shift to high glucose use and aerobic glycolysis (as is observed in cancers, wound healing, and immunological responses), and so responses to stress signals and control of these metabolic pathways is an important element of development. Abnormal cell division signaling during development in the retinoblastoma pathway (excessive E2F transcription factors) or the Ras or myc pathways result in the transcription of the ARF gene. The ARF protein binds to the MDM-2 protein and inhibits its activity, raising p53 levels and shutting down cell division, initiating senescence or apoptosis (Bates et al. 1996; de Stanchina et al. 1998; Palmero et al. 1998; Honda and Yasuda 1999). In this way, abnormal development of the embryo is blocked by the p53 pathway. Indeed, p53‐regulated gene products can shut down a number of developmental pathways: (1) SIAH is a p53‐regulated gene that lowers the levels of β catenin in WNT pathway signaling (Fiucci et al. 2004; Iwai et al. 2004); (2) the ratio of p53 and GLI, the transcription factor in the Hedgehog signaling pathway, regulates neural stem cell renewal (Stecca et al. 2009); and (3) the cell fate determinant NUMB activates p53 and inhibits the notch pathway cell determination (Carter and Vousden 2008; Colaluca et al. 2008). It is clear from these and other examples that the p53, 63, and 73 pathway genes play a role in integrating developmental steps with stress signals, and, in vertebrates, guiding several developmental pathways in the production of organisms.
CONCLUSIONS
The p63/73‐like gene was first observed in early metazoans such as the sea anemone (and possibly even single cell organisms such as the choanoflagellates), in which the function of the gene product was to protect the germ cells from environmental damage and stress, especially genomic instability. This function is so central to all organisms that it has been preserved, using the p53 family of genes, and extended over one billion years of evolution. Whether studied in detail in sea anemones, clams, fruit flies, round worms, zebrafish, mice, or humans, the p53, p63, and p73 genes appear to retain these functions, whereas in higher organisms, they take on additional attributes. With the advent of sexual dimorphism in a species, the p63 and p73 genes appear to play more important roles in the female germ line than the male germ line. In addition, some of the p53 family‐member‐regulated genes themselves have developed sexually dimorphic roles; i.e., LIF, WIP-1, and MDM-2 are estrogen and p53‐regulated genes and PERP is a progesterone and p53/63‐regulated gene that produces a tetraspanin protein important for skin cell adhesion (Attardi et al. 2000; Ihrie et al. 2005; Hu et al. 2007a; Hu et al. 2007b). In addition to germ‐line surveillance by the three sister genes, p53 has taken on the role of regulating the production of LIF in the uterus to regulate the implantation of the embryo. This has been directly observed in mice, and SNPs in the p53, MDM-2, MDM-4, and HAUSP, and LIF genes in humans can affect implantation in Caucasians (Kang et al. 2009). These functions ensure that some alleles in these genes will be under positive and negative evolutionary selection pressures. This, in turn, will have an impact upon development and cancer incidence and outcome. Remarkably, the functional role of germ‐line surveillance has been intact for more than one billion years.
The first change in the function of the p63/p73 ancestral gene coincided with the development of stem cells and progenitor cells designed to regenerate somatic tissues over the life time of the adult organism. As with germ cells, this created the possibility of unlimited cell growth, or cancers. At the evolutionary times in which this happened and in which we have information about the p53 family of genes (vertebrates), a duplication of the parent p63/p73 hybrid gene arose, creating the p53 gene, which evolved functions of somatic stem cell surveillance of genomic instability and other stress responses. This is what made the p53 gene a tumor suppressor gene. After this (the bony fish representative), the p63 and p73 genes were duplicated (the second major change in the family) and evolved some distinct developmental functions. From the production of functional germ cells, through the regulation of the implantation of embryos into the uterus in mammals, through development of the fetus into a newborn, the p53, p63, and p73 proteins function at various times and places to produce a normal organism. Throughout the lifetime of the vertebrate organism, one or more of these three genes continue to function in tissue regeneration and monitoring stem cell and progenitor cell fidelity. There is some evidence that implicates this family of genes in helping to set longevity of the organism or species (Donehower and Levine, 2008) and in implementing menopause as part of a homoeostatic mechanism (observed in worms and mice) (Rashidi 2008; Tavernarakis et al. 2008; Scrable et al. 2009). There is good evidence that the frequency of mutations and mistakes in genome segregation increases with age in both males and females. The efficiency of the p53‐mediated surveillance of these mistakes declines with age (Feng et al. 2008). It may well be that the surveillance and the prevention of these errors falls upon the p53/p63/p73 family of genes.
The first indication of MDM-2 as the major negative regulator of p53 is observed in vertebrates. In fish through humans, so much of the regulatory circuits of p53 go through the MDM-2 protein (AKT-1, AMP kinase, ARF, cyclin D-PP2A, etc.) (Harris and Levine 2005), that there must have been a sizable reorganization of p53, p63/p73 regulation in the vertebrates. In worms, the AKT kinase negatively regulates the p53 ancestor directly, whereas in vertebrates, the AKT kinase acts through MDM-2 to regulate p53 (Zhou et al. 2001; Gottlieb et al. 2002; Quevedo et al. 2007). In worms, the ABL-1 protein regulates the p63/73 gene after DNA damage, and in vertebrates, DNA damage activates the ABL kinase, which is known to phosphorylate the p73 protein, which in turn is capable of initiating apoptosis (Agami et al. 1999; Gong et al. 1999; Yuan et al. 1999; Deng et al. 2004; Mantovani et al. 2004). The study of these genes permits us to learn a great deal about the strategies employed during evolutionary changes that permit critical proteins to be smoothly regulated during the rewiring of regulatory circuits. We need to choose several interesting organisms in the evolutionary tree and uncover their regulatory circuits for the p53 family of genes.
Clearly, this description of the evolutionary events in the p53 family of genes depends upon a selective and incomplete record. The description of evolutionary changes in genes and their functions is therefore speculative and subject to interpretation of what we know at this time. It does help that the p53 family of genes and their functions are the subject of over fifty thousand articles and studies. When one finds remarkable conservation of genes and functions over a billion years of evolution and in organisms as diverse as the sea anemone, flies, worms, clams, fish, mice, and humans, the discussion gains a good deal of credibility.
Many interesting questions remain: (1) How many independent times did the p53 gene arise from the p63/p73‐hybrid-ancestor gene and in what organisms? (2) Did this truly coincide with the development of somatic stem and progenitor cells in those organisms? (3) Where did the duplication of the p63/p73 gene occur and how did the striking role of p63 in epithelial cell development arise? (4) What are the functional roles of p73 in the immune and nervous systems? Does p53 play a role in the central nervous system as well, and if so, what is it? (5) How do the functions of p53, p63, and p73 interface with the major signal transduction pathways that control the development of organisms? What is the extent to which p53/p63/p73 regulate metabolic pathways in development, cancer, and in the adult organism exposed to stress signals? (6) Are p63 and p73 true tumor suppressor genes in humans, and if so, under what conditions do they act to accomplish this? (7) What are the selective pressures acting upon different alleles in the p53, p63, and p73 genes in different human racial groups or ethnic groups? (8) What is the role of p53, p63, and p73 in the longevity of an organism? And many more interesting questions will surely be formulated and studied. What has become clear from this discussion is that the three sisters in the p53 family of genes have been shaped by evolutionary forces just as these gene products themselves have played a key role in shaping the evolution of organisms by imparting germ‐line fidelity and slowing variation among offspring. Those who play a role in ensuring the passage of information with few errors from generation to generation shape and guide the path that evolution takes. This is yet another reason why we need to know just how all of the genes in the pathways of these three sisters function in many diverse organisms.
After this article was completed and in print, a paper by Lane et al. (Lane et al. 2009) was brought to our attention. This manuscript identified p53 (38% identity to human) and MDM-2 (22% identity to human) proteins in the placozoans, whose origins and position in invertebrate evolution are under some debate. This brings up the possibility that p53 and MDM-2 genes are present in some invertebrates such as clams and placozoans and p63/p73 genes, with no detectable p53 or MDM-2 genes observed in other invertebrates (flies with 25% identity to p63/p73 in humans and worms with 15% identity to p63/p73 in humans). Some of these three genes, p53, p63, and p73, could have been lost, inter-converted, or gained during the evolution of the invertebrates. The closer relationships between p63 and p73 when compared to p53 and the more extensive evolutionary changes in the p53 gene that occurred in vertebrates suggest similar events (duplications, losses, or inter-conversions) in the vertebrates. Additional information will be required to determine if there are two separate lines of evolution (p53/MDM-2 or p63/p73 ancestor genes) in the invertebrates, one of which may have led to the vertebrates.
Footnotes
Several species, such as lancelet and sea squirts, also have tentative intronless paralogs; these could be pseudogenes or candidate p53 family genes that are unlikely predecessors for mammalian p53-family and are not shown in the tree. One possible scenario is that they arose through independent duplication events with intron loss (Mourier and Jeffares 2003)
Editors: Arnold J. Levine and David P. Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Agami R, Blandino G, Oren M, Shaul Y 1999. Interaction of c-Abl and p73α and their collaboration to induce apoptosis. Nature 399:809–813 [DOI] [PubMed] [Google Scholar]
- Arum O, Johnson TE 2007. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci 62:951–959 [DOI] [PubMed] [Google Scholar]
- Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T 2000. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev 14:704–718 [PMC free article] [PubMed] [Google Scholar]
- Bates S, Rowan S, Vousden KH 1996. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13:1103–1109 [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126:107–120 [DOI] [PubMed] [Google Scholar]
- Bottger S, Jerszyk E, Low B, Walker C 2008. Genotoxic stress-induced expression of p53 and apoptosis in leukemic clam hemocytes with cytoplasmically sequestered p53. Cancer Res 68:777–782 [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19:2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103–113 [DOI] [PubMed] [Google Scholar]
- Carter S, Vousden KH 2008. A role for Numb in p53 stabilization. Genome Biol 9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, et al. 1999. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143–153 [DOI] [PubMed] [Google Scholar]
- Chang J, Kim DH, Lee SW, Choi KY, Sung YC 1995. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem 270:25014–25019 [DOI] [PubMed] [Google Scholar]
- Cheng R, Ford BL, O’Neal PE, Mathews CZ, Bradford CS, Thongtan T, Barnes DW, Hendricks JD, Bailey GS 1997. Zebrafish (Danio rerio) p53 tumor suppressor gene: cDNA sequence and expression during embryogenesis. Mol Mar Biol Biotechnol 6:88–97 [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP 2008. NUMB controls p53 tumour suppressor activity. Nature 451:76–80 [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93:519–529 [DOI] [PubMed] [Google Scholar]
- Cox RL, Stephens RE, Reinisch CL 2003. p63/73 homologues in surf clam: Novel signaling motifs and implications for control of expression. Gene 320:49–58 [DOI] [PubMed] [Google Scholar]
- de Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev 12:2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hofmann ER, Villanueva A, Hobert O, Capodieci P, Veach DR, Yin X, Campodonico L, Glekas A, Cordon-Cardo C, et al. 2004. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat Genet 36:906–912 [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH 2001. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 294:591–595 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Levine AJ 2008. p53, cancer and longevity, In Molecular Biology of Aging, (eds) Guarente Leonard, Partridge Linda and Wallace Douglas, Cold Spring Harbor Laboratory Press, 6, 127–152 [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1:45–49 [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ 2007. The regulation of AMPK β1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 67:3043–3053 [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci 102:8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Rajagopal G, Levine AJ 2008. The tumor suppressor p53: Cancer and aging. Cell Cycle 7:842–847 [DOI] [PubMed] [Google Scholar]
- Fiucci G, Beaucourt S, Duflaut D, Lespagnol A, Stumptner-Cuvelette P, Geant A, Buchwalter G, Tuynder M, Susini L, Lassalle JM, et al. 2004. Siah-1b is a direct transcriptional target of p53: Identification of the functional p53 responsive element in the siah-1b promoter. Proc Natl Acad Sci 101:3510–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T 2005. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7:363–373 [DOI] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG Jr, Levrero M, Wang JY 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806–809 [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Leal JF, Seger R, Taya Y, Oren M 2002. Cross-talk between Akt, p53 and Mdm2: Possible implications for the regulation of apoptosis. Oncogene 21:1299–1303 [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ 2005. The p53 pathway: Positive and negative feedback loops. Oncogene 24:2899–2908 [DOI] [PubMed] [Google Scholar]
- Holbrook LA, Butler RA, Cashon RE, Van Beneden RJ 2009. Soft-shell clam (Mya arenaria) p53: A structural and functional comparison to human p53. Gene 433:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 18:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YJ, Wang DY, Cheng CC, Chen JK 2004. Age-related expressions of p63 and other keratinocyte stem cell markers in rat cornea. J Biomed Sci 11:641–651 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Atwal GS, Levine AJ 2008. p53: A new player in reproduction. Cell Cycle 7:848–852 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Ma L, Wagner J, Rice JJ, Stolovitzky G, Levine AJ 2007a. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res 67:2757–2765 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ 2007b. p53 regulates maternal reproduction through LIF. Nature 450:721–724 [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. 2009. Ensembl 2009. Nucl Acids Res 37:D690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Jeffrey PD, Derry WB, Rothman JH, Pavletich NP, Stavridi ES, Halazonetis TD 2004. Structural differences in the DNA binding domains of human p53 and its C. elegans ortholog Cep-1. Structure 12:1237–1243 [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD 2005. Perp is a p63-regulated gene essential for epithelial integrity. Cell 120:843–856 [DOI] [PubMed] [Google Scholar]
- Iwai A, Marusawa H, Matsuzawa S, Fukushima T, Hijikata M, Reed JC, Shimotohno K, Chiba T 2004. Siah-1L, a novel transcript variant belonging to the human Siah family of proteins, regulates β-catenin activity in a p53-dependent manner. Oncogene 23:7593–7600 [DOI] [PubMed] [Google Scholar]
- Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, Mills AA, Miller FD, Kaplan DR 2005. p63 is an essential proapoptotic protein during neural development. Neuron 48:743–756 [DOI] [PubMed] [Google Scholar]
- Jayaraman J, Prives C 1995. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81:1021–1029 [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267:1498–1502 [DOI] [PubMed] [Google Scholar]
- Jessen-Eller K, Kreiling JA, Begley GS, Steele ME, Walker CW, Stephens RE, Reinisch CL 2002. A new invertebrate member of the p53 gene family is developmentally expressed and responds to polychlorinated biphenyls. Environ Health Perspect 110:377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci 97:7301–7306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Feng Z, Sun Y, Atwal JK, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine A, Hu W 2009. Single nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci 106:9761–9766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucl Acids Res 36:D773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley ML, Winge P, Heaney JD, Stephens RE, Farell JH, Van Beneden RJ, Reinisch CL, Lesser MP, Walker CW 2001. Expression of homologues for p53 and p73 in the softshell clam (Mya arenaria), a naturally-occurring model for human cancer. Oncogene 20:748–758 [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z 2006. Structural basis of DNA recognition by p53 tetramers. Mol Cell 22:741–753 [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948–953 [DOI] [PubMed] [Google Scholar]
- Lane DP, Cheak CF, Brown C, Madhumalar A, Ghadessy F, Verma C 2009. MDM-2 and p53 are highly conserved from placozoans to man. Cell Cycle (in press). [DOI] [PubMed] [Google Scholar]
- Le Goas F, May P, Ronco P, Caron de Fromentel C 1997. cDNA cloning and immunological characterization of rabbit p53. Gene 185:169–173 [DOI] [PubMed] [Google Scholar]
- Lee H, Kimelman D 2002. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell 2:607–616 [DOI] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J 26:4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Feng Z, Mak TW, You H, Jin S 2006. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev 20:267–275 [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ 1994. Several hydrophobic amino acids in the amino-terminal domain are required for transcriptional activation, binding to MDM-4 and Adenovirus E1B55Kd. Genes Develop 8:1235–1246 [DOI] [PubMed] [Google Scholar]
- Lu WJ, Abrams JM 2006. Lessons from p53 in non-mammalian models. Cell Death Differ 13:909–912 [DOI] [PubMed] [Google Scholar]
- Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, Blandino G, Del Sal G 2004. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell 14:625–636 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM 2006. p53 regulates mitochondrial respiration. Science 312:1650–1653 [DOI] [PubMed] [Google Scholar]
- Melino G, Lu X, Gasco M, Crook T, Knight RA 2003. Functional regulation of p73 and p63: Development and cancer. Trends Biochem Sci 28:663–670 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708–713 [DOI] [PubMed] [Google Scholar]
- Moll UM, Slade N 2004. p63 and p73: Roles in development and tumor formation. Mol Cancer Res 2:371–386 [PubMed] [Google Scholar]
- Mourier T, Jeffares DC 2003. Eukaryotic intron loss. Science 300:1393. [DOI] [PubMed] [Google Scholar]
- Nahor I, Abramovitch S, Engeland K, Werner H 2005. The p53-family members p63 and p73 inhibit insulin-like growth factor-I receptor gene expression in colon cancer cells. Growth Horm IGF Res 15:388–396 [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Tan C 2007. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol 217:801–806 [DOI] [PubMed] [Google Scholar]
- Norimura T, Nomoto S, Katsuki M, Gondo Y, Kondo S 1996. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med 2:577–580 [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91–101 [DOI] [PubMed] [Google Scholar]
- Ortt K, Sinha S 2006. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS letters 580:4544–4550 [DOI] [PubMed] [Google Scholar]
- Ou H, Lohr F, Vogel V, Mantele W, Dotsch V 2007. Structural evolution of C-terminal domains in the p53 family. EMBO J 26:3463–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125–126 [DOI] [PubMed] [Google Scholar]
- Pan H, Dung HN, Hsu HM, Hsiao KM, Chen LY 2003. Cloning and developmental expression of p73 cDNA in zebrafish. Biochem Biophys Res Commun 307:395–400 [DOI] [PubMed] [Google Scholar]
- Pankow S, Bamberger C 2007. The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. PLoS ONE 2:e782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park US, Su JJ, Ban KC, Qin L, Lee EH, Lee YI 2000. Mutations in the p53 tumor suppressor gene in tree shrew hepatocellular carcinoma associated with hepatitis B virus infection and intake of aflatoxin B1. Gene 251:73–80 [DOI] [PubMed] [Google Scholar]
- Pietsch EC, Sykes SM, McMahon SB, Murphy ME 2008. The p53 family and programmed cell death. Oncogene 27:6507–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintus SS, Fomin ES, Oshurkov IS, Ivanisenko VA 2007. Phylogenetic analysis of the p53 and p63/p73 gene families. In Silico Biol 7:319–332 [PubMed] [Google Scholar]
- Quevedo C, Kaplan DR, Derry WB 2007. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr Biol 17:286–292 [DOI] [PubMed] [Google Scholar]
- Rashidi A 2008. On evolution of menopause. Maturitas 59:283–284 [DOI] [PubMed] [Google Scholar]
- Rippin TM, Freund SM, Veprintsev DB, Fersht AR 2002. Recognition of DNA by p53 core domain and location of intermolecular contacts of cooperative binding. J Mol Biol 319:351–358 [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. 2009. Database resources of the National Center for Biotechnology Information. Nucl Acids Res 37:D5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol 11:1722–1727 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S 2005. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 12:153–161 [DOI] [PubMed] [Google Scholar]
- Scrable H, Medrano S, Ungewitter E 2009. Running on empty: How p53 controls INS/IGF signaling and affects life span. Exp Gerontol 44:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siah A, Delaporte M, Pariseau J, McKenna P, Berthe FCJ 2009. Patterns of p53, p73 and mortalin gene expression associated with haemocyte polyploidy in the soft-shell clam, Mya arenaria. J Inverterb Pathol 98:148–152 [DOI] [PubMed] [Google Scholar]
- Signoretti S, Pires MM, Lindauer M, Horner JW, Grisanzio C, Dhar S, Majumder P, McKeon F, Kantoff PW, Sellers WR, et al. 2005. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci 102:11355–11360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M 2008. Characterization of genome-wide p53-binding sites upon stress response. Nucl Acids Res 36:3639–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame N, Kim M, Abrams JM 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci 100:4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Bègue A, Kress M, Stehelin D, May P 1988. Nucleotide sequence of a cDNA encoding the chicken p53 nuclear oncoprotein. Nucleic Acids Res 16:11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz I Altaba A 2009. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J 28:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F 2006. p63 protects the female germ line during meiotic arrest. Nature 444:624–628 [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G 2008. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 4:870–873 [DOI] [PubMed] [Google Scholar]
- Teufel DP, Freund SM, Bycroft M, Fersht AR 2007. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci 104:7009–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. 2008. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22:2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoglu M, Karaca E, Haliloglu T, Nussinov R 2008. Cataloging and organizing p73 interactions in cell cycle arrest and apoptosis. Nucl Acids Res 36:5033–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, et al. 2001. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet 69:481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veprintsev DB, Freund SM, Andreeva A, Rutledge SE, Tidow H, Canadillas JM, Blair CM, Fersht AR 2006. Core domain interactions in full-length p53 in solution. Proc Natl Acad Sci 103:2115–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, et al. 2006. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J 25:5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KK, Levine AJ 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci 93:15335–15340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Farmer G, Soussi T, Prives C 1995. Xenopus laevis p53 protein: Sequence-specific DNA binding, transcriptional regulation and oligomerization are evolutionarily conserved. Oncogene 10:779–784 [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F 1998. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2:305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714–718 [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99–103 [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D 1999. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399:814–817 [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 3:973–982 [DOI] [PubMed] [Google Scholar]