Figure 2.
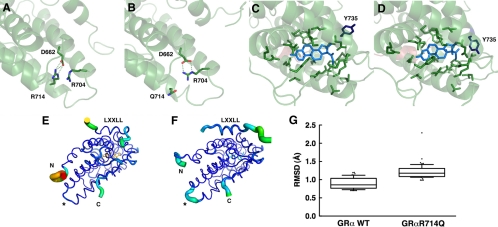
Arginine (R) to glutamine (Q) replacement at amino acid 714 of the hGRα alters three-dimensional structure of its LBD. A and B, Arginine (R) to glutamine (Q) replacement at amino acid 714 of the hGRα causes formation of a new salt bridge between arginine (R) at 704 and aspartic acid (D) at 662. In the wild-type hGRα LBD, R714 is tightly bound to D662 through an electrostatic salt bridge (A). Substitution of arginine for glutamine in the hGRαR714Q LBD results in a rearrangement of the side chains, forming a new salt bridge between R704 and D662 and displacing Q714 (B). This relaxes some constraint on helix 10 and results in structural changes throughout the LBD. C and D, hGRαR714Q LBD has altered ligand-binding pocket. Ligand-binding pockets of the wild-type hGRα (C) and hGRαR714Q (D) are shown. Arginine to glutamine replacement at amino acid 714 causes alteration in the ligand-binding pocket in hGRαR714Q LBD. The greatest change observed between the wild-type hGRα LBD and hGRαR714Q LBD is in the rotametric state of tyrosine (Y) 735. E–G, hGRαR714Q LBD has the destabilized AF-2 surface that blocks optimal binding of the LXXLL coactivator motif. The thickness and color of the Cα trace of the wild-type hGRα LBD (E) and hGRαR714Q LBD (F) indicate the areas of least (thin and blue) to most (thick and red) motion over the course of the simulation. As expected, the termini of the LBD and the bound peptide move the most. There is significant motion at the mutation site (*) in the mutant LBD (F) but not in the wild-type, suggesting the mutation caused destabilization in this structural area. The bound LXXLL peptide is thus more labile in the mutant LBD structure as seen by the thickness of its Cα trace and quantified by the box plots (bars indicate 10th and 90th percentiles, and dots show outlined results) in G, in which the median root mean square deviation (RMSD) for the helical residues containing the LXXLL motif (amino acids 743–750 of the human transcription intermediary factor 2) is 1.2 Å with the mutant LBD over the course of the simulation, whereas it is only 0.86 Å with the wild-type LBD (P < 0.0001), suggesting that the mutant structure binds the peptide with less affinity.