Abstract
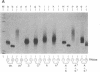
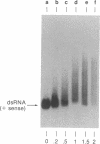
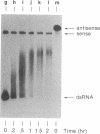
Amphibian eggs and embryos as well as mammalian cells have been reported to contain an activity that unwinds double-stranded RNA. We have now found that adenosine residues have been modified in the RNA products of this unwinding activity. Although the modified RNA remains double-stranded, the modification causes the RNA to be susceptible to single-strand-specific RNase and to migrate as a retarded smear on a native polyacrylamide electrophoresis gel. The modification is specific for double-stranded RNA. At least 40% of the adenosine residues can be modified in vitro in a given random sequence RNA molecule. By using standard two-dimensional TLC and HPLC analyses, the modified base has been identified as inosine. Mismatched base-pairing between inosine and uridine appears to be responsible for the observed characteristics of the unwound RNA. The biological significance of this modifying activity and also of the modified double-stranded RNA is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987 Feb 27;48(4):607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Buck M., Connick M., Ames B. N. Complete analysis of tRNA-modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal Biochem. 1983 Feb 15;129(1):1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- Calabretta B., Robberson D. L., Maizel A. L., Saunders G. F. mRNA in human cells contains sequences complementary to the Alu family of repeated DNA. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6003–6007. doi: 10.1073/pnas.78.10.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Schmid A., Eschle D., Baczko K., ter Meulen V., Billeter M. A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988 Oct 21;55(2):255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., Weng S. A., Silberman S. R., Cai S. J., Deslypere J. P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987 Oct 16;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Feagin J. E., Abraham J. M., Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988 May 6;53(3):413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981 Jan 15;145(2):405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rebagliati M. R., Melton D. A. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987 Feb 27;48(4):599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- Richter J. D., Smith L. D., Anderson D. M., Davidson E. H. Interspersed poly(A) RNAs of amphibian oocytes are not translatable. J Mol Biol. 1984 Feb 25;173(2):227–241. doi: 10.1016/0022-2836(84)90191-8. [DOI] [PubMed] [Google Scholar]
- Shaw J. M., Feagin J. E., Stuart K., Simpson L. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988 May 6;53(3):401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Gietz R. D., Hodgetts R. B. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986 Jul 17;322(6076):279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Wagner R. W., Nishikura K. Cell cycle expression of RNA duplex unwindase activity in mammalian cells. Mol Cell Biol. 1988 Feb;8(2):770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]