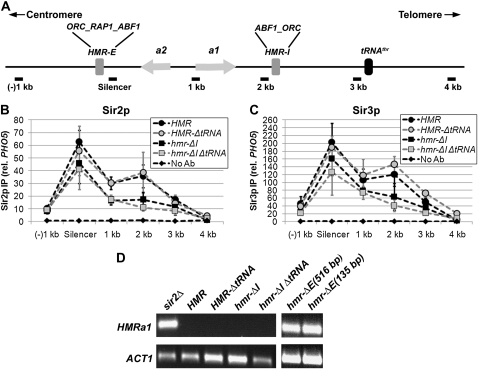
Figure 3.—
The HMR-I silencer, but not the tRNAThr gene, boosts Sir protein levels within HMR. (A) Diagram of the wild-type HMR locus. (B) Association of Sir2p with HMR in the presence and absence of regulatory elements. Sir2p-associated DNA was isolated by chromatin IP from strains with the following genotypes: wild-type HMR (LRY1007), HMR-ΔtRNAThr (LRY2302), hmr-ΔI (LRY2315), and hmr-ΔI ΔtRNAThr (LRY2309). Data were analyzed as in Figure 1B and represent the averages of at least three independent immunoprecipitation experiments, each analyzed in duplicate quantitative PCR reactions. Primer sets and relative distances were the same for each mutant, with the exception that the +4-kb location is 85 bases closer to the silencer in strains lacking the tRNAThr gene. (C) Association of Sir3p with HMR in the presence and absence of regulatory elements. Sir3p-associated DNA was isolated and analyzed as in B. (D) Transcription of HMRa1 in the presence and absence of regulatory elements. RNA was isolated from the same strains described above, a sir2Δ strain with wild-type HMR (LRY1068) and two different strains with large (YAB65, 516 bp) and small (YAB71, 135 bp) deletions of the HMR-E silencer. The mRNA transcripts were converted to cDNA and the resulting cDNA was amplified by conventional PCR using primer sequences specific to the coding regions of HMRa1 and ACT1. The PCR products were run on 1% agarose gels and visualized by ethidium bromide staining.