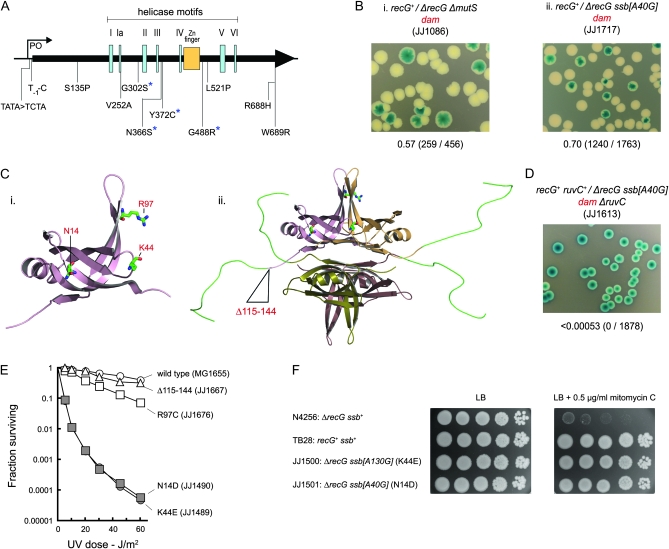
Figure 5.—
Suppressors of the low viability of dam recG cells. (A) Map showing location and identity of priA suppressors. Coding mutations are indicated in terms of the changes to PriA. The asterisks identify changes that provide for particularly strong suppression of the mitomycin C sensitivity of a recG strain. (B) Synthetic lethality assays illustrating restoration of viability to recG dam cells by mutS and ssb mutations. (C) Structure of an SSB monomer (i) and tetramer (ii) showing the amino acid residues affected by the ssb mutations identified. The models were generated using Pymol and SSB crystal coordinates from the PDB database (Matsumoto et al. 2000; Raghunathan et al. 2000; Savvides et al. 2004). (D) Synthetic lethality assay showing that the viability conferred on recG dam cells by ssb[A40G] depends on RuvC. (E) Effect of ssb mutations on sensitivity to UV light. (F) Restoration of mitomycin C resistance to a recG strain by ssb[A40G] and ssb[A130G]. Cultures of the strains indicated were grown in LB broth to an A650 of 0.4, diluted in 10-fold steps, and 10-μl aliquots spotted on LB agar without and with mitomycin C, as indicated. The plates were photographed after 24 hr at 37°.