Table 2.
PDE isoform selectivity data for 1, 5, 10 and 18.
| PDE isof orm* |
1 I C50 /% inh. |
5 I C50 /% inh. |
10 I C50 /% inh. |
18 I C50 /% inh. |
|
|---|---|---|---|---|---|
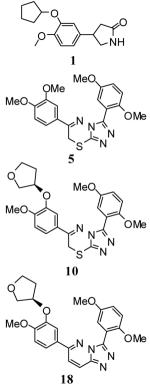
|
PDE1A | inactive | inactive | 36% | 32% |
| PDE1B | inactive | inactive | 52% | 56% | |
| PDE1C | inactive | 26% | 49% | 74% | |
| PDE2A | inactive | 41% | 68% | 54% | |
| PDE3A | inactive | 1.7 μM | 56% | 54% | |
| PDE3B | inactive | 720 nM | 4.6 μM | 2.3 μM | |
| PDE4A1A | 102 nM | 12.9 nM | 0.26 nM | 0.6 nM | |
| PDE4B1 | 901 nM | 48.2 nM | 2.3 nM | 4.1 nM | |
| PDE4B2 | 534 nM | 37.2 nM | 1.6 nM | 2.9 nM | |
| PDE4C1 | 40% | 452 nM | 46 nM | 106 nM | |
| PDE4D2 | 403 nM | 49.2 nM | 1.9 nM | 2.1 nM | |
| PDE5A1 | inactive | 60% | 58% | 51% | |
| PDE7A | inactive | 73% | 48% | 59% | |
| PDE7B | inactive | 33% | 43% | 35% | |
| PDE8A1 | inactive | 57% | inactive | inactive | |
| PDE9A2 | inactive | inactive | inactive | inactive | |
| PDE10A1 | inactive | 823 nM | 632 nM | 388 nM | |
| PDE11A4 | inactive | inactive | inactive | inactive |
data generated by BPS Biosciences (CA) [http://www.bpsbioscience.com]and represents either the IC50 value or the % inhibition at 10 μM of compound.
