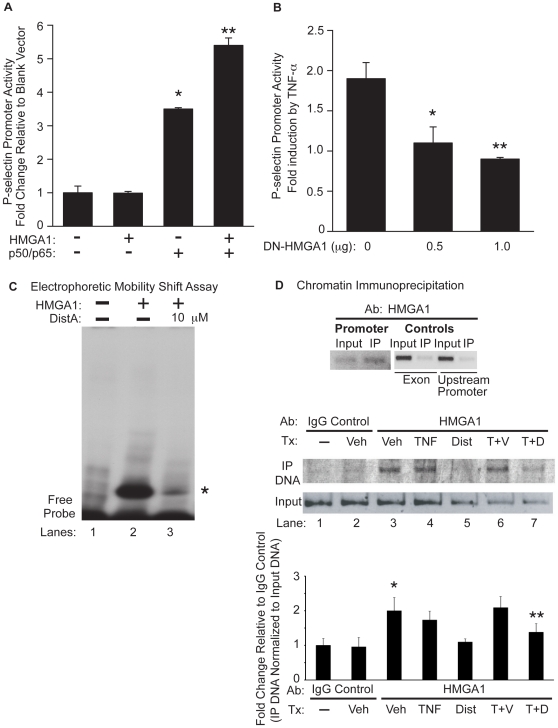
Figure 6. HMGA1 binds to the P-selectin promoter and is critical for full induction of P-selectin promoter activity.
A. BAEC cells were transiently transfected with a P-selectin promoter-reporter construct with the addition of a blank expression vector, an expression vector for HMGA1, and/or expression vectors for NF-κB family members (p50/p65). Transfected cells were harvested and assessed for luciferase activity (normalized for β-galactosidase content). Results are expressed as fold-change in luciferase activity relative to transfection with the P-selectin promoter and a blank expression vector. B. BAEC cells were transiently transfected with a P-selectin promoter-reporter construct and increasing concentrations of a vector expressing a dominant-negative form of HMGA1 (DN-HMGA1). Transfected cells were stimulated with TNF-α, then harvested and assessed for luciferase activity (normalized for β-galactosidase content). Results are expressed for each transfection condition as fold-change in luciferase activity as a result of TNF-α stimulation. (*p<0.05 for 0.5 µg of DN-HMGA1 as compared with empty vector control; **p<0.05 for 1.0 µg of DN-HMGA1 as compared with empty vector control). This experiment was repeated three separate times, with each condition performed in triplicate. C. An electrophoretic mobility shift assay (EMSA) was performed using the HMGA1 peptide and a radiolabeled probe spanning the AT-rich region of the P-selectin promoter (basepairs −542 to −521) without (Lane 2) or with Dist A (10 µM, Lane 3). Lane 1 represents the radiolabeled probe in the absence of incubation with protein. (* represents the HMGA1-DNA complex in Lane 2 which is diminished in intensity following addition of Dist A in Lane 3). Binding studies were repeated at least two separate times. D. Chromatin Immunoprecipitation (ChIP) was performed on murine bEnd.3 endothelial cells without (Lane 1) or with 3 hours of exposure to Vehicle (Veh, Upper Panel and Lanes 2–3 Lower Panel), mTNF-α (TNF, Lane 4), Distamycin A (Dist A, Lane 5), mTNF-α/Vehicle (T+V, Lane 6), and mTNF-α/Dist A (T+D, Lane 7). Immunoprecipitation of crosslinked, sonicated cell lysates was carried out with an affinity-purified HMGA1 antibody (Upper panel and Lanes 3–7 Lower Panel) or IgG rabbit control antibody (Lanes 1–2 Lower Panel), with PCR amplification using primers spanning a 246-basepair AT-rich region of the P-selectin promoter (including basepairs −542 to −521; “Promoter” in Upper Panel and all lanes in Lower Panel) or 200-basepair fragments spanning a coding region or upstream promoter region of P-selectin (“Exon” and “Upstream Promoter”, respectively in Upper Panel; see sequences in Methods section). PCR amplification was undertaken for immunoprecipitated DNA as well as for “input” DNA as an additional control for each condition. Quantitation of precipitated DNA relative to input DNA was undertaken using Quantity One 1-D Analysis Software (Bio-Rad). This experiment was performed two separate times. (*p<0.05, for increase in binding for Veh (HMGA1 Ab) compared with IgG control; **p<0.05, for reduction in binding for TNF-α/Dist compared with TNF-α/Vehicle).