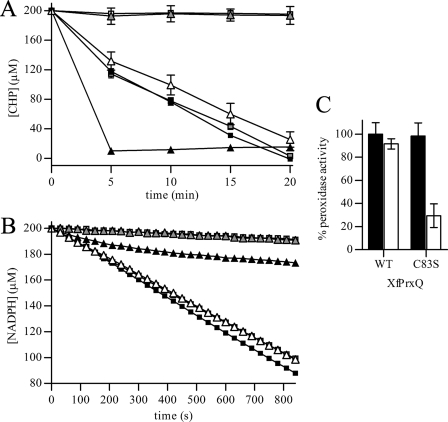
FIGURE 3.
Effects of the Cys → Ser mutations on XfPrxQ disulfide bond formation and peroxidase activity. A, removal of CHP in DTT-dependent peroxidase assay by XfPrxQ proteins as follows: wild-type (gray squares), XfPrxQ C23S (black squares), XfPrxQ C47S (gray triangles), XfPrxQ C83S (black triangles), and XfPrxQ C101S (open triangles). Peroxidase reactions were carried out in a reaction mixture containing 200 μm CHP, 2 mm DTT, and 25 μm XfPrxQ proteins at 37 °C. As a negative control, a peroxidase reaction was performed without XfPrxQ protein (open squares). At the indicated times, the remaining CHP was quantified in triplicate, and results are the mean ± S.D. (error bars). B, NADPH oxidation coupled to CHP reduction in the presence of XfPrxQ proteins as follows: wild-type (gray squares), XfPrxQ C23S (black squares), XfPrxQ C47S (gray triangles), XfPrxQ C83S (black triangles), and XfPrxQ C101S (open triangles). Assays were carried out in a reaction mixture containing 250 μm CHP, 200 μm NADPH, 1 μm XfPrxQ proteins, 1 μm E. coli Trx, and 0.1 μm E. coli TrxR at 37 °C. As a negative control, peroxidase reactions were performed without XfPrxQ protein (open squares). C, dimedone inactivation of XfPrxQ wild-type and C83S proteins. Prior to the DTT-dependent peroxidase assay, XfPrxQ proteins were incubated with 1.1 eq of hydrogen peroxide in the absence (control, black bars) or presence of 1,000 eq of dimedone (white bars) at room temperature for 30 min. Peroxidase reactions were carried out in a mixture containing 300 μm TBHP, 1 mm DTT, 1 μm E. coli Trx, and 10 μm XfPrxQ proteins at 37 °C. After 10 min, the remaining TBHP was quantified in triplicate, and results are the mean ± S.D. (error bars). Peroxidase activity was considered 100% when wild-type XfPrxQ was assayed in the absence of dimedone.