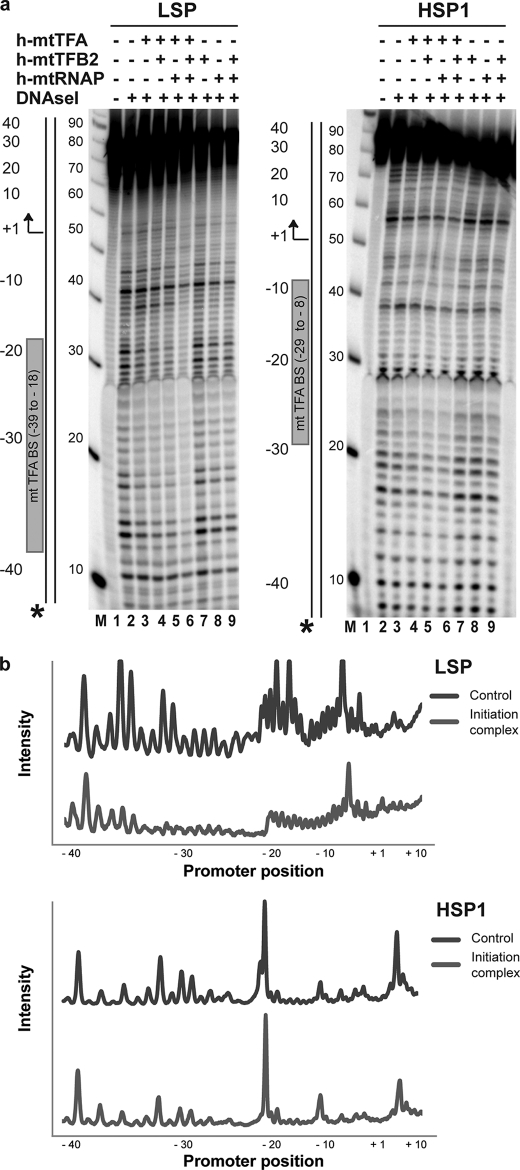
FIGURE 4.
DNase I footprinting provides evidence for assembly of initiation complexes on LSP and HSP1 oligo templates. DNase I footprinting was performed using LSP and HSP1 DNA oligonucleotide templates. Proteins were assembled on the indicated 32P-labeled DNA oligo (2.5 μm) by combining one or more of the following components: h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm), as indicated, in reaction buffer at 32 °C. DNA cleavage was initiated by addition of RQ1 DNase (0.002 unit/μl) and CaCl2 (1 mm) and quenched after 2 min by addition of stop/trap buffer. Products were resolved by denaturing PAGE on 8% gels and visualized by phosphorimaging. The locations/orientations of the h-mtTFA-binding site and transcription start site are indicated for the LSP and HSP1 promoters. A 10-bp ladder was used as a size marker (M). a, footprinting was performed on LSP and HSP1 containing 32P in the 5′ end of the templating strand. b, quantification of selected lanes of the gels shown in a. The intensities were normalized to the −40 position of the control (lane 2 of LSP or HSP1 in a). Initiation complex refers to lane 6 of LSP or HSP1 in a. Asterisk refers to the location of the 32P label.