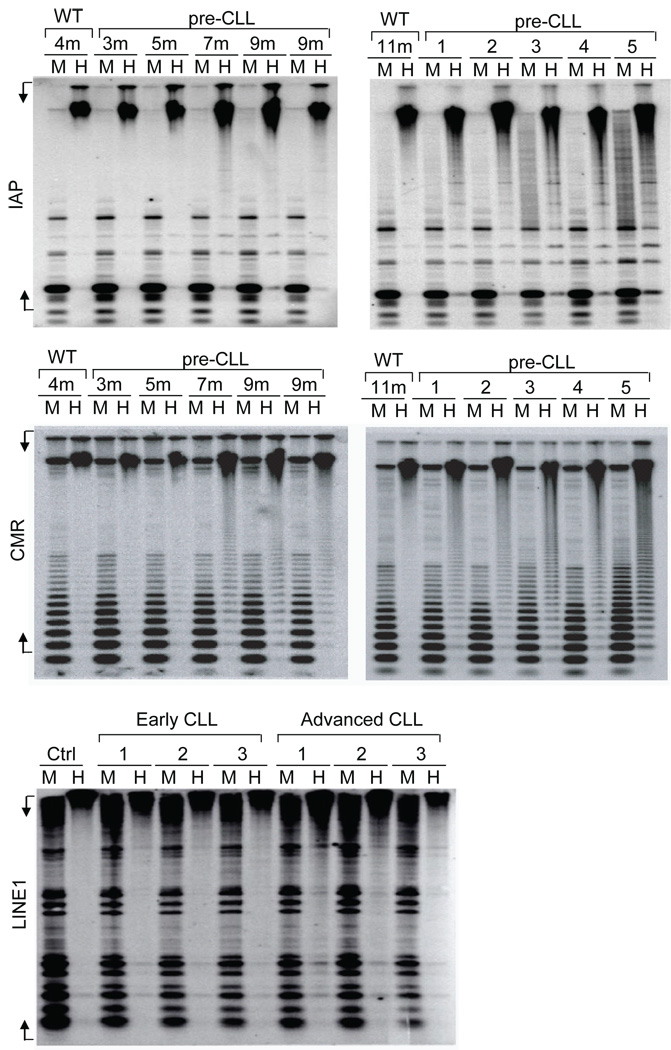
Figure 1. Elevated global hypomethylation of repetitive sequences in CLL.
Hypomethylation profiles on mouse and human CLL samples from different stages were studied by Southern blotting as previous described34. Genomic DNAs of spleen cells from wild type and Eµ-TCL1 mice at indicated time points, or Eµ-TCL1 mice with symptomatic disease (n=5), were digested with MspI (M) or HpaII (H) restriction enzymes. The blots were hybridized with intracisternal A particle probe (IAP) (Top) or centromeric repeat sequences (CMR) (Middle). For CLL patient samples, the analysis was done using human LINE1 probes on the peripheral blood B cells from patients required no treatment (early CLL) or that underwent treatment (advanced CLL) (Bottom). MspI digested DNA was used as control. Undigested product of HpaII digestion represents methylated DNA, while only unmethylated DNA is HpaII digestible. Mouse IAP, CMS and human LINE1 probes were prepared by PCR amplification using forward (F) and reverse (R) primers: IAP-F: 5’CGTCATTGTTCAGAGCCAGA3’, IAP-R: 5’TCCCGGAAACTTTTGTTCAC3’; CMS-F: 5’GATAAAAACCTACACTGTAG3’, CMS-R: 5’GTTTCTAATTGTAACTCATTG3’; LINE1-F: CGGGTGATTTCTGCATTTCC and LINE1-R: GACATTTAAGTCTGCAGAGG.