Table 1.
In Vitro Data for Quinolines, Benzothiazoles and MPEP or MTEP-like Alkynyl mGluR5 Antagonistsa
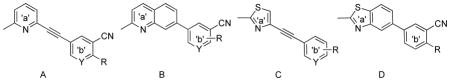 | ||||||
|---|---|---|---|---|---|---|
| Compound ID | template | Y | R | mGluR5 binding affinity Ki±SEM(nM) | mGluR5 function(Ca+2 flux) IC50±SEM(nM) | clogPb |
| 1, MPEP | A | CH | H, 3-Hc | 13±1d | 3.54±1.39d | 3.8 |
| 2, MTEP | C | N | H, 3-Hc | 16e | 13.6±2.09d | 2.1 |
| 3 | B | CH | H | 110±20f | 29±5f | 3.9 |
| 4 | D | H | 2100±580f | NT | 3.9 | |
| 6 | A | CH | H | 1.3±0.09d | 0.415±0.10d | 3.2 |
| 7 | C | CH | 3-CN-5-F | 0.9±0.2d | 0.813±0.11d | 3.2 |
| 8a | A | CH | Ph | 4.0±0.6d | 3.08±0.61d | 5.1 |
| 8b | A | CH | 4 -F-Ph | 3.0±0.5d | 7.19±1.53d | 5.3 |
| 8c | C | N | 4-Ph | 5.49±1.43d | 1.21± 1.15d | 4.2 |
| 8d | C | CH | 3-F-4-3 -Py | 11.4±3d | 3.43±0.51d | 3.0 |
| 10 | B | CH | 4-OH | >10,000 | >10,000 | 3.9 |
| 13 | D | OTf | 862±200 | 7840±1250 | 5.4 | |
| 14 | D | 4-F-Ph | 4679±1185 | >10,000 | 5.9 | |
| 16 | B | CH | 4-OCH2OCH3 | 596±109 | 294+21 | 3.6 |
| 17 | B | CH | 4-OBz | 1040±206 | >10,000 | 5.9 |
| 18 | D | OBz | 7134±2026 | >10,000 | 5.9 | |
| 21 | B | CH | 3-Hc-2-CN-4-Cl | 4720±913 | 9150±1540 | 4.6 |
| 22 | B | CH | 4-Ph | 97±20 | 1250±261 | 5.7 |
| 23 | B | CH | 4-4 -F-Ph | 64±16 | 692±64 | 5.9 |
| 24 | B | CH | 4-3 -Py | 730±190 | 1340±41 | 4.3 |
| 25 | D | Ph | 483±98 | 6890±1250 | 5.8 | |
| 26 | D | 3-Py | >10,000 | >10,000 | 4.3 | |
| 28 | A | N | H | 1.5±0.3 | 13±2.3 | 1.9 |
| 30g | B | N | H | 100±21 | 68±5.3 | 2.5 |
| 31h | B | N | 4-Cl | >10,000 | 3310±384 | 3.2 |
| 32 | B | N | 4-Ph | 97±19 | 81±8.1 | 4.7 |
Methods for binding (http://pdsp.med.unc.edu/pdspw/binding.php) and functional assays19 have been previously published;
Determined with ChemDraw Ultra 10.0;
3-H, no CN group on the 3-position of structure;
Data reported previously in literature.19;
Data reported previously in literature.25;
Data reported previously in literature.16;
Compound reported previously in literature14;
Partial antagonist; NT = not tested.
