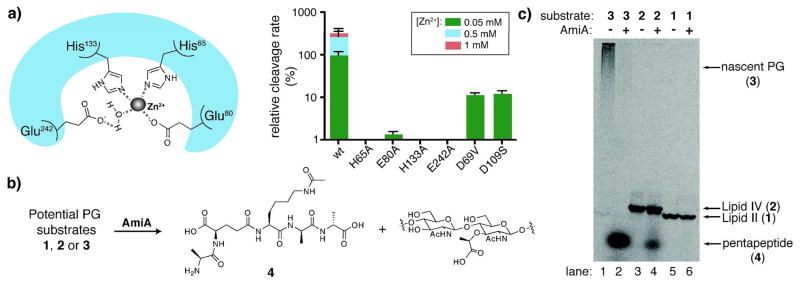
Figure 2.
Analysis of AmiA substrate preferences and catalytic features: (a) predicted zinc-binding site of AmiA based on alignment with B. polymyxa var. colistinus CwlV (left); comparison of relative cleavage rates of 3 (7.2 μM) by mutants of AmiA (4.0 μM) (right); (b) reaction scheme for AmiA cleavage of [14C]-pentapeptide (4) from potential PG substrates that differ in length; (c) gel electrophoresis of 3 (lanes 1, 2), 2 (lanes 3, 4), and 1 (lanes 5, 6) without and with AmiA addition, respectively, under similar reaction conditions. AmiA cleaves 3 and 2, but not 1, to produce a new band that represents 4.