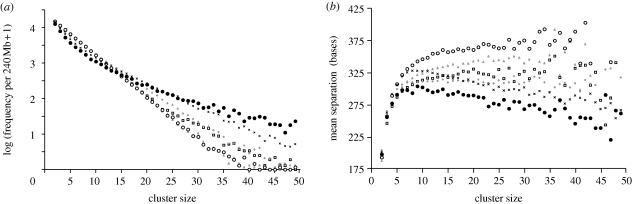
Figure 1.
How the frequencies and densities of SNP clusters vary with cluster size under different mutation models. Data series are: large black circles, data from human chromosome 1; large white circles, simulated randomly occurring mutations; smaller symbols, simulated mutation non-independence in which mutations are more likely to occur in regions of size 0.5 (grey triangles), 1 (white squares), 2 (grey diamonds) or 5 kb (crosses) around any existing heterozygous site. Simulated data are culled from 100 replicate simulations, accepting only those in which the terminal overall SNP density was one SNP every 700 ± 25 bases, yielding approximately 60 runs per set of conditions. Frequencies are normalized to the size of human chromosome 1. For full details of the simulations (see §2). (a) How the frequencies of different cluster sizes vary; (b) how cluster density varies for the same data.