Fig. 2.
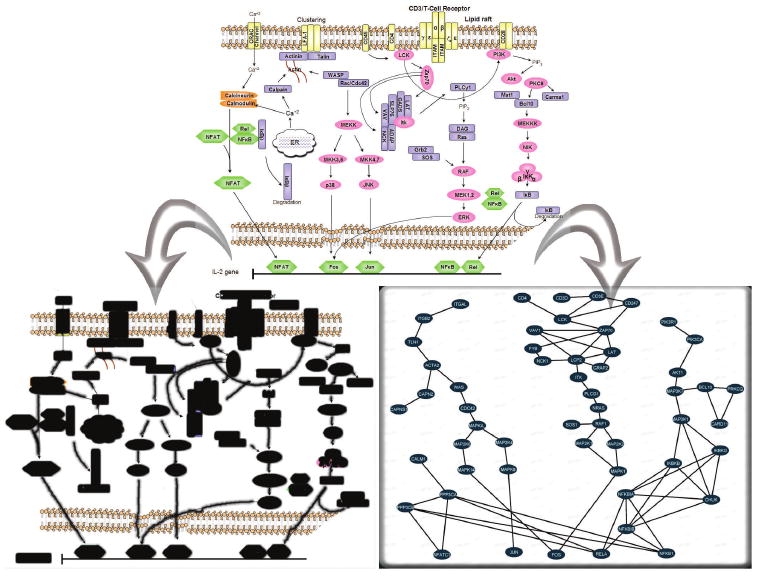
Structuring protein interactions around familiar canonical pathways provides intuitive visualizations. A canonical signaling pathway representation (top) can be imported into the system in two ways: on the lower left the pathway image itself is loaded into the system and preprocessed by circling proteins and drawing over interactions; the pathway features are then inferred from the user strokes and image features and shown here in black; or, on the lower right, protein and interaction icons are placed and dragged on an empty canvas to create a new pathway model. After positional assignment of each protein, the software aids in associating interaction database accession numbers to each of the newly defined canonical pathway proteins.
