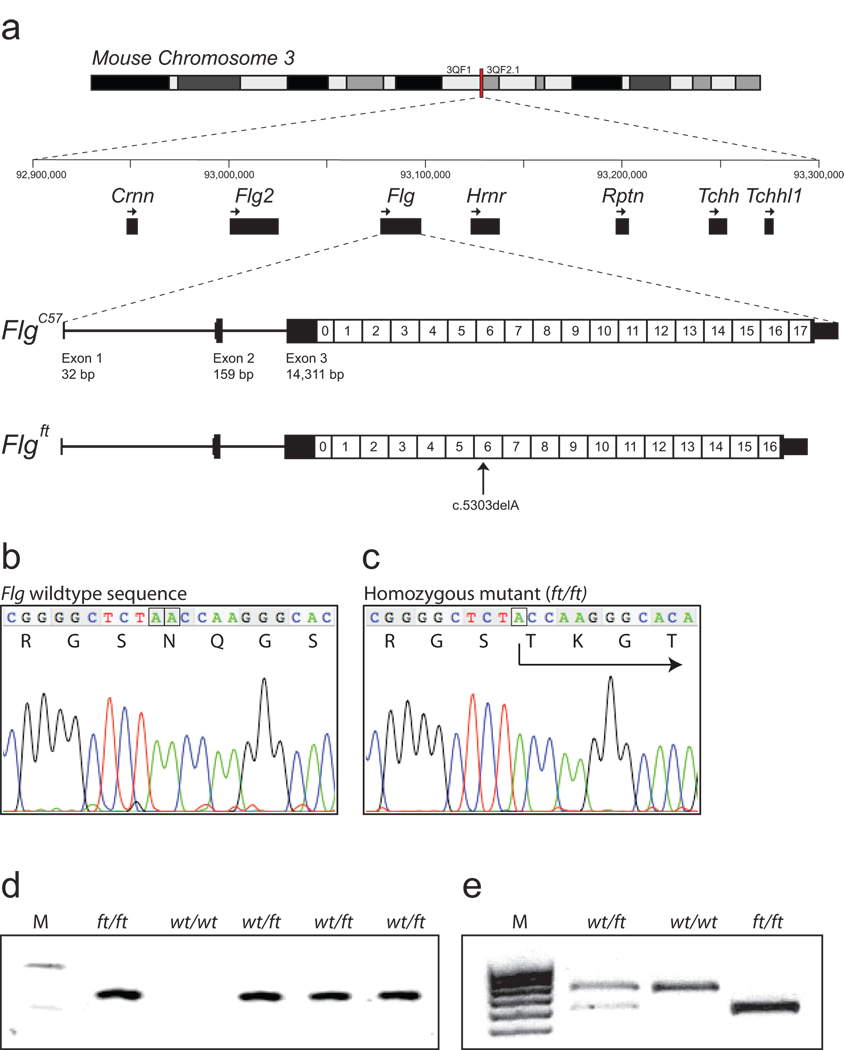
Figure 1. Murine filaggrin gene structure and mutation identification.
(a) The mouse filaggrin gene (flg) resides within a cluster of 7 genes encoding closely related fused-S100 proteins, spanning ~350 kb on murine chromosomal band 3QF21. Like the human ortholog, flg has a 3 exon gene structure. Exon 1 consists of 5’UTR sequences only; exon 2 contains the ATG and encodes part of the N-terminal S100 calcium binding domain of profilaggrin; and exon 3 encodes the remainder of the S100 domain, the B-domain and the filaggrin polyprotein repeat domain. In the July 2007 mouse genome sequence, annotated here (FlgC57), the latter consists of 16 near-perfect repeats encoding a 250 amino acid filaggrin protein (1–16) flanked by two imperfect repeats (0 and 17). Repeat 17 is followed by a short tail domain. The 485 bp 3’UTR contains a consensus polyadenylation signal. In ft mice, the exon 3 sequence determined here (Flgft) lacks one repeat due to a 750-bp in-frame deletion, as well as the frameshift mutation c.5303delA. In addition, there are several additional SNPs and small in-frame insertions/deletions in flg that differ between the two strains, so that their DNA sequences overall are 99.3% identical, excluding the repeat copy number variation.
(b) Wild-type flg sequence, corresponding to codons 1765–1771, with amino acid translation. The A dinucleotide involved in the mutation is boxed (compare panel c).
(c) Analogous flg sequence as shown in (b), derived from a homozygous ft/ft mouse, showing homozygous frameshift mutation designated c.5303delA, predicting truncation mutation p.Asn1768ThrfsX154. The single A nucleotide remaining at the site of the mutation is boxed (compare panel b).
(d) Allele-specific genotyping of the parents and F1 offspring of a cross between an ft/ft homozygote and a wild-type mouse. M, molecular weight markers, upper band = 118 bp; lower band = 72 bp. The allele specific band is only amplified from mice carrying the ft mutation.
(e) Genotyping for an Acc I restriction site polymorphism in filaggrin repeat 1, approximately 4 kb upstream of the 5303delA mutation. This assay is more convenient for scoring of heterozygotes. The 678 bp C57BL/6 allele does not digest; the ft allele cuts to yield fragments of 559 bp and 134 bp. M, molecular weight markers.