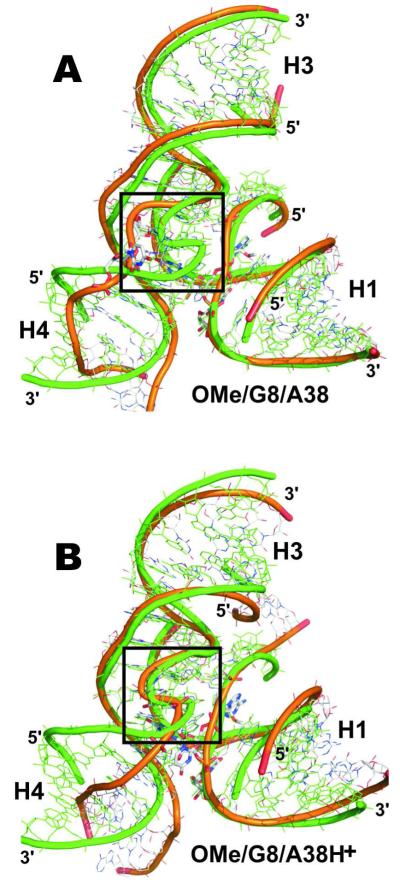
Figure 6.
Ribbon diagrams showing the average structures from the last ns (orange ribbon) superimposed over the crystal structure (green) of the minimal junction-less hairpin ribozyme with helixes H1-H4 indicated. (A) Simulations with canonical A38 (here OMe/G8/A38) show S-turn degradation (black box). (B) Simulations with protonated A38H+ (here OMe/G8/A38H+) preserve the crystallographic S-turn conformation.