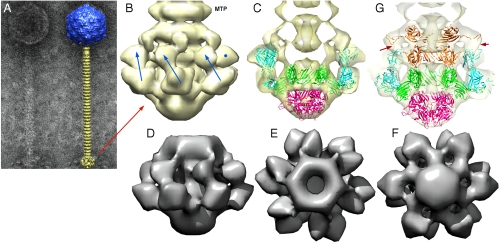
Fig. 2.
EM structure of the baseplate of phage p2 and results of the fitting of the baseplate crystal structure. (A) EM micrograph of the virulent lactococcal phage p2. Overall EM structure of phage p2 (right) showing the capsid (blue, top), the tail, formed of rings of the major tail protein hexamers (gold), and the globular baseplate (gold, bottom). ( B) View of the EM structure of the baseplate with the first major tail protein (MTP) hexamer. The RBP positions are identified by blue arrows or a blue dot. The RBP head is pointing upwards. (C) View of the crystal structure of the baseplate fitted in the EM map. ORF15 is green, ORF16 red, and ORF18 blue. Each RBP, ORF15 hexamer, and ORF16 trimer have been fitted as rigid blocks. (D), (E), (F) Views of the cryoEM map of the expressed p2 baseplate, from side (D), up (E), and down (F), respectively. (G) Sliced view showing the fitting of a second ORF15 hexamer (brown) in the EM map, 180° rotated relative to that in the crystal structure. The ring formed by the N-terminus has been fitted as a block, and the rotated and translated galectin domains have been fitted as a second block. Note the extension touching the RBP head domain (red arrows).