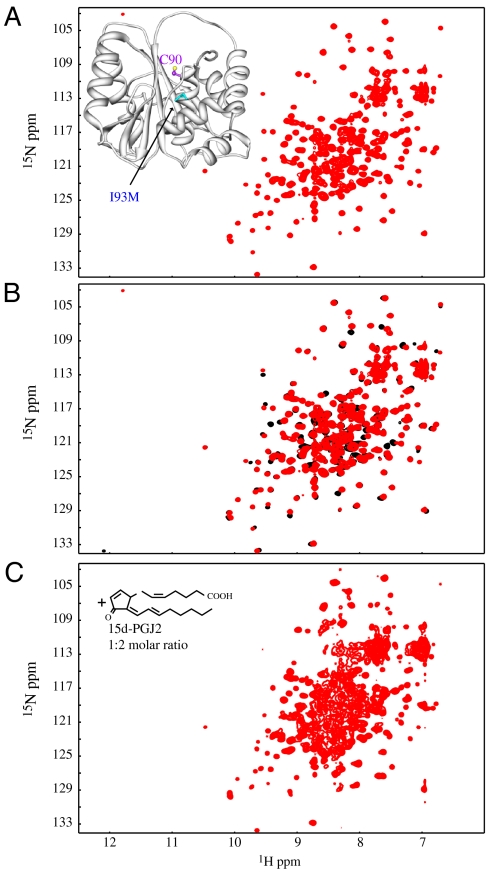
Fig. 4.
The structure of the I93M mutant is similar to wild-type protein and undergoes 15d-PGJ2 modification, unfolding and aggregation. (A) 2D 1H-15N HSQC spectrum of the UCH-L1 I93M mutant exhibits excellent chemical shift dispersion, demonstrating a well-folded protein. In the inset a ribbon of the x-ray structure (9) is displayed, with the location of the mutated residue marked in cyan. (B) Superposition of the 2D 1H-15N HSQC spectra of wild-type UCH-L1 (Black) and the I93M mutant (Red). Only very small changes in resonance positions were observed, indicating that no significant structural changes between I93M mutant and wild-type proteins are caused by the mutation. (C) 2D 1H-15N HSQC spectrum of the I93M mutant in the presence of 2 fold molar excess of 15d-PGJ2. All spectra were recorded for 0.2 mM protein samples in PBS buffer (pH 7.6) at 298 K.