Abstract
MyTH/FERM (myosin tail homology 4/band 4.1, ezrin, radixin, and moesin) myosins have roles in cellular adhesion, extension of actin-filled projections such as filopodia and stereocilia, and directional migration. The amoeba Dictyostelium discoideum expresses a simple complement of MyTH/FERM myosins, a class VII (M7) myosin required for cell-substrate adhesion and a unique myosin named MyoG. Mutants lacking MyoG exhibit a wide range of normal actin-based behaviors, including chemotaxis to folic acid, but have a striking defect in polarization and chemotaxis to cAMP. Although the myoG mutants respond to cAMP stimulation by increasing persistence and weakly increasing levels of cortical F-actin, they do not polarize; instead, they maintain a round shape and move slowly and randomly when exposed to a chemotactic gradient. The mutants also fail to activate and localize PI3K to the membrane closest to the source of chemoattractant. These data reveal a role for a MyTH/FERM myosin in mediating early chemotactic signaling and suggest that MyTH/FERM proteins have conserved roles in signaling and the generation of cell polarity.
Keywords: actin cytoskeleton, cell signaling, cell motility, cytoskeletal dynamics
The directed movement of cells is essential for multicellular development and the ability of organisms to fight off infection (1, 2). Cellular translocation is driven by dynamic rearrangements of the actin cytoskeleton and its associated motors. Pseudopod extension occurs via localized actin polymerization at the plasma membrane, and forward movement of the cell body depends upon actomyosin-based cortical tension in the rear. Cell-surface adhesion receptors linked to the underlying actin cytoskeleton provide mechanical integration of the cell and its substrate. Members of the myosin superfamily of actin-based motor proteins contribute to several different aspects of cell migration (3). For example, filamentous myosin II (M2) has a critical role in determining cell polarity. The cortical localization of M2 at the rear of the cell is required for the normal preferential distribution of cytoplasm toward the anterior portion of the cell and the formation of a tapered uropod. In contrast, myosin I (M1) family members regulate the timing and location of pseudopod formation in amoeba, most likely by delivering or localizing components of the actin polymerization machinery to the plasma membrane at the site of pseudopod extension.
A group of myosins, referred to as the MyTH/FERM myosins, have been implicated in cell-substrate adhesion and have roles in the extension of specialized actin-based projections, such as filopodia and stereocilia (3). These myosins are characterized by the presence of both MyTH4 (myosin tail homology 4) and FERM (band 4.1, ezrin, radixin, and moesin) domains, typically adjacent to each other in their C-terminal tail regions. MyTH4 domains bind to microtubules and are found in diverse proteins, including plant kinesins (4, 5). FERM domains bind to membrane receptors and are characteristically found in proteins that play a role in linking cell-surface adhesion receptors to the actin cytoskeleton or regulating the interactions between adhesion proteins (6). The class VII (M7), X (M10), and XV (M15) myosins are MyTH/FERM myosins. M7a contributes to cadherin-based adhesion in the sensory hair cells of the vertebrate ear, where it interacts via its C-terminal FERM domain with adapter proteins that bind the cyotplasmic tails of cadherins (7). M7a may also play dual roles in adhesion, by both trafficking adhesion-receptor complexes to their correct cellular location and providing a link to the underlying cytoskeleton. M10 binds to integrins via its FERM domain and is thought to traffic them to the cell periphery and then provide a stable link between the actin cytoskeleton and substrate-bound integrin (8). The FERM domain of Drosophila M15 binds to DE-cadherin, and M15 is also thought to transport the cadherin along filopodia for deposition at sites of cell-cell adhesion (9). MyTH/FERM myosins also regulate actin dynamics. Depletion of Dictyostelium M7 and vertebrate M10 from cells results in a loss of filopodia and M15-deficient sensory hair cells of the ear extend short stereocilia (10–12). Interestingly, the sensory hair cells in mice lacking M7a extend longer stereocilia (13), suggesting that this myosin plays a role in negatively regulating growth of these actin-based structures. Two MyTH/FERM myosins have been directly implicated in the transport of regulators of actin polymerization to the tips of filopodia and stereocilia. The tail of M10 interacts with Mena/VASP and transports VASP to the tips of filopodia (14, 15) and the M15 tail binds to the PDZ protein whirlin, localizing it to the tips of stereocilia, where it promotes stereocilia growth (16).
The social amoeba Dicytostelium expresses two MyTH/FERM myosins, M7 and MyoG, a myosin with a tail domain structure strikingly similar to that of M7 (Fig. 1A) (17), consisting of a tandem repeat of MyTH/FERM domains separated by an SH3 domain. However, phylogenetic analysis reveals that MyoG is not a member of the known classes of MyTH/FERM myosins and it is currently considered to be an orphan myosin. The possibility that MyTH/FERM myosins have a fundamentally conserved role in cellular adhesion and the regulation of actin dynamics, or that they may have a wider variety of functions than previously appreciated, was investigated through the generation and analysis of a myoG null mutant.
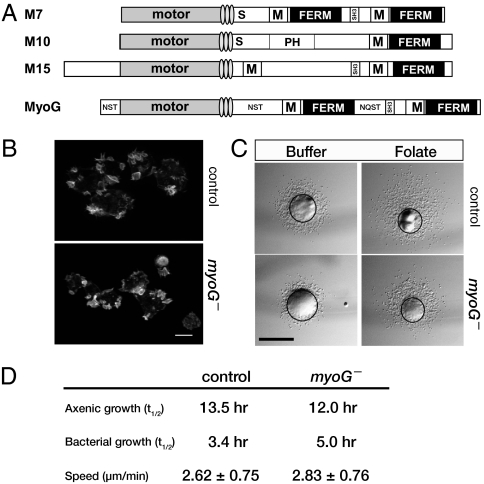
Fig. 1.
MyoG does not have a general role in actin-based functions. (A) Domain organization of several different MyTH/FERM myosins. The locations of the light-chain binding IQ motifs (lightly shaded ovals), single alpha-helix region (S), MyTH4 (M), and pleckstrin-homology domains (PH) are indicated. The MyoG heavy chain contains several regions enriched for the amino acids asparagine (N), serine (S), threonine (T), and glutamate (Q) and the location and major amino acids are indicated. (B) Confocal projections of vegetative control and myoG null mutants fixed and stained for actin. (Scale bar, 10 μm.) (C) Representative examples of the small-drop folate chemotaxis assay at 2 h. The well of either buffer or folate is toward the top of each panel. The black circle overlay indicates the circumference of the spot at the 0 h time-point. (Scale bar, 1 mm.) (D) Rates of growth and motility for control and myoG null mutants. Shown are the average doubling times (t1/2) of control and myoG null mutants in nutrient broth (axenic) or in B/r bacteria. Shown are the averaged results from two independent experiments (n ≥ 4). The speed (mean ± SD) of vegetative cells was determined by averaging the results of three independent experiments (total n ≥ 58 cells per strain).
Results
MyoG is a unique large MyTH/FERM myosin (predicted molecular weight, ∼387 kDa) that is phylogenetically distinct from unconventional myosins, such as M7, M10, and M15, which have similar tail domain structures (Fig. 1A) (17). The cellular function of MyoG was investigated through the generation and analysis of mutants carrying a disrupted myoG gene (Fig. S1 A and B). Two independently generated mutant strains (HTD36-2 and HTD36-5) were analyzed in detail and found to behave identically in all assays. For simplicity, only results from the HTD36-2 line are presented here. The phenotype of the myoG mutants was compared to either the parental Ax2 cells or control nonhomologous recombinants (NHR) that behaved identically to Ax2 cells. The potential role of MyoG in general cytoskeletal function was assessed first by measuring several distinct actin-based behaviors of vegetative myoG mutant cells. The organization of the actin cytoskeleton in the myoG mutants is indistinguishable from that of control cells. The mutant cells possess normal cytoskeletal structures, including filopodia and macropinocytic crowns (Fig. 1B). A small-drop assay (18) was performed to measure the ability of the myoG mutants to move and chemotax toward folic acid. Spots of cells placed around a well of folate were scored for net movement toward chemoattractant. A total of 95% of the wild-type spots and 90% of myoG null spots (n = 30) showed positive chemotaxis to folic acid, compared with spots placed around a well of buffer that exhibited unbiased movement outwards from the spot (Fig. 1C). Growth in liquid medium, a measure of the ability of cells to internalize fluids as well as carry out cytokinesis, and basal cell motility of myoG mutants are both similar to that of control cells (Fig. 1D). However, the rate of growth in bacterial suspension, a process that depends on phagocytic activity, is somewhat slower than that of wild-type cells (an average of 5 h vs. 3.4 h for the parental wild-type cells), indicating that the mutants may have a mild phagocytosis defect. The behavior of the mutant cells in these assays suggests that MyoG is not required for overall actin cytoskeletal function but that it may contribute to phagocytosis.
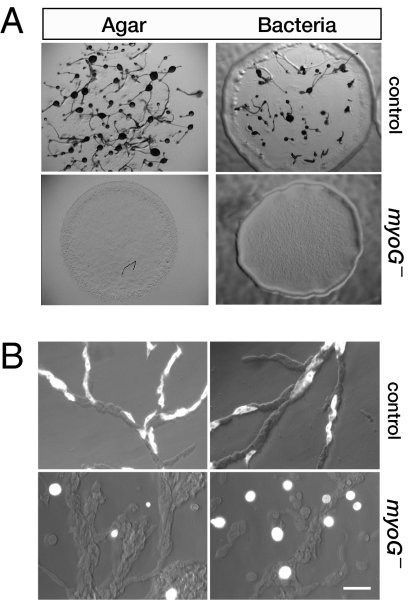
The removal of nutrients from Dictyostelium triggers a developmental program that relies on the ability of cells to chemotax to cAMP and results in the formation of a fruiting body consisting of a slender stalk topped by a round head full of spores. The myoG mutants exhibit a complete failure to develop in response to starvation under all conditions tested (Fig. 2A). Wild-type cells deposited on nonnutrient agar formed fruiting bodies after 24 h, whereas the myoG nulls failed to develop, arresting as smooth lawns that lack any multicellular structures, even after 48 h. Similarly, myoG mutant cells spotted on a bacterial lawn formed plaques that expanded over time, but these were smooth and never showed signs of development, in contrast to the control cells that formed fruiting bodies in the center of the expanding plaque (Fig. 2A). The early developmental defect is consistent with the expression of myoG in both vegetative and starving cells (Fig. S1C). Aggregation-competent Dictyostelium are highly polarized and move rapidly, coming together to form streams. Control and myoG mutant cells expressing coronin-GFP were mixed with unlabeled Ax2 cells to assess the behavior of these cells during aggregation. The control cells are highly polarized and fully incorporated into streams; the myoG null cells appear quite round and rarely participate in streaming (Fig. 2B). These results reveal that the myoG mutant developmental defect is cell autonomous and suggests that the mutants either fail to enter the developmental program or have a chemotaxis defect.
Fig. 2.
MyoG is required for aggregation. (A) The development of control and myoG null cells was assayed by placing a concentrated drop of cells (>1 × 108 cell/mL) on starvation agar or by spotting a small number of cells on a lawn of B/r bacteria. Samples were photographed at 24 h or 3 days, respectively. The control cells form typical fruiting bodies consisting of a slender stalk topped by a spore head; the mutants arrest as a smooth lawn. (B) Mixing experiment illustrating the failure of the myoG null cells to polarize and move toward aggregation centers in streams. A mixture of coronin-GFP expressing control or myoG null cells with untagged control cells (15% GFP cells and 85% nonfluorescent cells) was pulsed with cAMP, allowed to adhere to a coverslip after 6 h, and streams observed shortly thereafter. The differential interference contrast and fluorescent images were overlaid to illustrate the location and morphology of the coronin-GFP expressing cells. (Scale bar, 20 μm.)
The initiation of the developmental program and chemotaxis both rely on the ability of cells to detect and respond to factors secreted in to the environment (19). An early response to starvation is the expression of two key chemotaxis genes: one encoding the cAMP receptor, cAR1 (carA), and the other the adenylyl cyclase responsible for cAMP production (acaA) (20). The failure to express these key early genes could account for the observed phenotype of the myoG null mutant, so their expression immediately following the removal of nutrients was analyzed by semiquantitative RT-PCR. Starving myoG mutant cells increase the expression of both carA and acaA, at the same time and to the same extent as control cells (Fig. S2A), indicating that the myoG mutant can detect and respond to the secreted signals that initiate the developmental program. Myosins have roles in intracellular trafficking (3); therefore, the possibility that the myoG mutants fail to develop because they are defective in trafficking of the key chemotaxis receptor, cAR1, to the cell surface was tested. A cAR1-GFP fusion was expressed in both control and myoG mutant cells and found to be localized to the plasma membrane in both cell types (Fig. S2B). Together, these data strongly suggest that the myoG mutant developmental defect is not the result of a failure to sense developmental cues or traffic the key chemotaxis receptor but is instead because of an inability to respond to the chemoattractant cAMP.
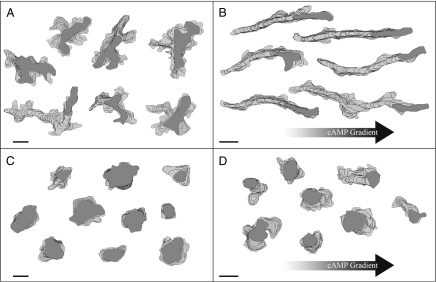
Chemotaxis to cAMP is characterized by a highly polarized cell shape and rapid, persistent movement toward the chemoattractant source (21). The myoG null mutants that fail to incorporate into streams (Fig. 2B) appear unpolarized and slow-moving. Therefore, a detailed quantitative analysis of aggregation-competent myoG mutants using time-lapse microscopy and the Dynamic Image Analysis System software (22) was performed to assess the response of cells to a chemotactic stimulus. Control and mutant cells were placed in either a buffer perfusion chamber or a cAMP gradient chamber and their behavior recorded and analyzed. Perimeter tracks of control cells in a buffer perfusion chamber (no signaling) show highly active cells producing multiple pseudopodia at random sites around the cell periphery (Fig. 3A). When placed in a gradient of cAMP, control cells polarize and move up the gradient at an average velocity of 6.4 ± 2.3 μm/min and with a chemotactic index of 0.54 ± 0.20 (Fig. 3B, Table 1). In contrast, myoG mutant cells in buffer have a smaller area and produce fewer extensions around the cell body (Fig. 3C, Table 1). This result is similar to what is observed for vegetative cells that also have a smaller area than controls (73.50 ± 37.94 vs. 101.35 ± 37.94 μm2; P = 3.97 E-08) and suggests that the mutants have a defect in cell spreading. The myoG null cells placed in a cAMP gradient are noticeably round, indicating that they fail to polarize (Fig. 3D, Table 1). The mutants also do not chemotax, having a chemotactic index of only 0.05 ± 0.22. Control cells in a cAMP gradient move persistently, translocating in one direction for a period. The persistence of wild-type cells almost triples when comparing cells in buffer alone to cells in a gradient (Fig. 3B, Table 1). Interestingly, the myoG null mutants also exhibit a noticeable increase in persistence when placed in a cAMP gradient (about 2-fold), although the direction of persistent movement is not oriented toward the cAMP source (Fig. 3D, Table 1). These results reveal that although the myoG null mutants do exhibit a mild response to chemoattractant, they are profoundly defective in both polarization and chemotaxis.
Fig. 3.
The myoG mutant cells fail to polarize or chemotax in response to cAMP. Cells were pulsed with cAMP for 6 h and placed in a buffer perfusion chamber or a cAMP gradient. Shown here are representative perimeter tracks of NHR control (A and B) and myoG mutant (C and D) cells in buffer (A and C) or a chemotactic gradient (B and D). The filled dark-gray cell perimeter represents the cell's position in the last frame of the movie. Perimeters shown are from every fifth frame (starting with frame 1) of a 10-min movie captured at 15 frames per min. (Scale bars, 10 μm.)
Table 1.
Quantitative analysis of the myoG mutant response to cAMP
| Assay condition | Strain | Number of cells | Instantaneous velocity (μm/min) | Directional persistence (net/total dist.) | Direction change (deg./4 s) | Positive flow (%/4 s) | Max. length (μm) | Max. width (μm) | Area (sq μm) | Roundness (%) | Chemotactic index (CI) | % cells positive CI |
| Buffer perfusion | Control | 37 | 3.9 ± 1.3 | 0.20 ± 0.18 | 48.25 ± 11.1 | 4.72 ± 0.9 | 30.7 ± 8.9 | 18.0 ± 6.6 | 304.03 ± 170.01 | 46.3 ± 13.4 | — | — |
| myoG- | 69 | 2.4 ± 0.9 | 0.12 ± 0.09 | 50.5 ± 12.0 | 3.41 ± 1.1 | 20.5 ± 7.3 | 14.5 ± 5.1 | 221.25 ± 151.96 | 79.4 ± 11.0 | — | — | |
| P value | 5.72E-08 | 1.49E-02 | NS | 7.07E-09 | 1.25E-07 | 6.43E-03 | 1.57E-02 | 3.11E-19 | — | — | ||
| cAMP gradient | Control | 33 | 6.4 ± 2.3 | 0.59 ± 0.19 | 38.3 ± 15.3 | 7.96 ± 2.7 | 23.7 ± 5.5 | 6.53 ± 1.1 | 95.87 ± 26.46 | 42.6 ± 9.3 | 0.54 ± 0.20 | 100 |
| myoG- | 57 | 2.4 ± 0.8 | 0.27 ± 0.17 | 35.2 ± 10.0 | 3.44 ± 0.8 | 16.3 ± 4.4 | 10.6 ± 2.1 | 119.78 ± 49.88 | 77.2 ± 10.9 | 0.05 ± 0.22 | 56 | |
| P value | 1.39E-11 | 3.96E-11 | 1.96E-12 | 9.63E-20 | 2.09E-08 | 2.78E-20 | 3.86E-03 | 7.31E-26 | 5.32E-17 | — |
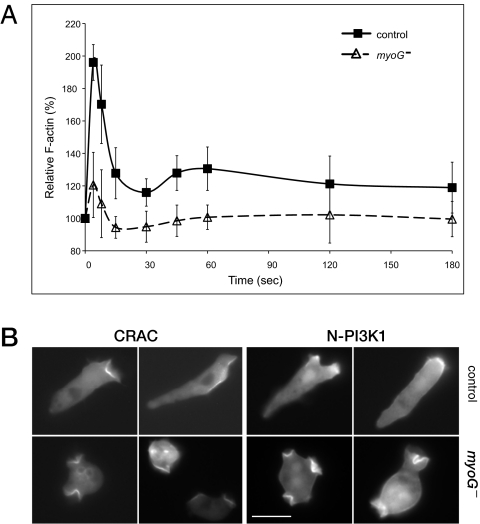
The acquisition of a polarized shape and directional movement toward a source of attractant results from a signal transduction cascade initiated by cAMP binding to cAR1 that promotes rapid alterations in the actin cytoskeleton. Stimulation with cAMP causes characteristic changes in the levels of cortical F-actin polymerization that are correlated with stereotypical transformations in cell shape (23, 24). An initial peak of increased cortical F-actin, about 2-fold, is observed ≈5 s after a cAMP pulse, resulting in rounding up of the cell. A second smaller, broader peak follows within 45 to 60 s after the cAMP pulse and this correlates with the extension of small pseudopodia all around the cell periphery. The ability of the myoG null cells to transiently increase cortical F-actin in response to cAMP was tested to determine if signaling to the cytoskeleton, which could account for the observed phenotype, was defective in these mutants. Stimulation of the myoG mutant cells with cAMP did result in a burst of actin polymerization at 5 s, but the overall response was significantly dampened (Fig. 4A). A 20% increase in cortical F-actin was observed as compared to the 100% increase seen for control cells. Furthermore, the second smaller and broader peak of actin polymerization at 40 to 60 s was not observed (Fig. 4B). Thus, the myoG null cells are severely impaired in signaling to the actin polymerization machinery in response to cAMP stimulation.
Fig. 4.
Chemotactic signaling is impaired in myoG null cells. (A) Actin polymerization in response to cAMP is dampened in myoG- cells. Analysis of changes in cytoskeletal F-actin levels in both control (■) and myoG mutant (△) cells after a 100-nM cAMP pulse (administered at time = 0). Data shown from three independent experiments represent the mean ± SD of control (n = 8) and myoG null cells (n = 14). (B) Localization of CRAC-GFP and N-PI3K1-GFP in a cAMP gradient. Aggregation competent cells were exposed to a cAMP gradient in a Zigmond chamber and photographed after ~15 min. The cAMP source is at the right in all panels. (Scale bar, 10 μm.)
Localized actin polymerization occurs following the activation of small G proteins and PI3K at the side of the cell closest to the chemoattractant source (21). Activated PI3K, in turn, produces local increases of PIP3, providing docking sites for PH-domain containing proteins like CRAC (cytosolic regulator of adenylate cyclase) (25–28).The early steps in signal transduction in the myoG null cells were investigated first by examining the distribution of CRAC-GFP in cells placed in a cAMP gradient. Wild-type cells in the gradient exhibited a polarized distribution of CRAC-GFP, which is concentrated on the membrane closest to the cAMP source (Fig. 4B). The myoG null cells failed to localize CRAC-GFP on the membrane exposed to the highest concentration of chemoattractant (Fig. 4B). Instead, CRAC-GFP is found in membrane protrusions randomly located around the cell periphery. This distribution is the same as that observed in both wild-type and myoG-null vegetative cells and is most likely because of random, signal-independent activation of the basal PI3K associated with the plasma membrane (29). Similarly, PI3K is not localized at the side of the cell closest to the gradient in the myoG null cells but it is present on randomly extended protrusions (Fig. 4B). A transient PI3K localization event could have been masked by incubation of myoG null cells in a stable gradient of cAMP before observation. PI3K membrane recruitment was therefore assayed in cells pulsed with a single, high concentration of cAMP. Within 6 s of a 100 nM cAMP pulse, CRAC-GFP moves from the cytosol to the cell cortex in control cells (Fig. S2C). Membrane association is transient, dissipating after ∼12 s In contrast, GFP-CRAC remains in the cytosol of myoG null cells following the cAMP pulse (Fig. S2C). These data show that although myoG null cells exhibit basal PI3K activity, it is not locally stimulated in response to cAMP signaling.
Discussion
The unconventional myosin MyoG plays a critical role in cell polarization and chemotaxis. The failure of both of these processes in mutants lacking myoG is neither a result of general cytoskeletal nor global chemotactic dysfunction (Fig. 1 B–D). Vegetative myoG mutants grow normally in liquid culture, move at rates similar to control cells, and exhibit a strong chemotactic response to folic acid. The failure of mutant cells to aggregate in response to starvation (Fig. 2) cannot be attributed to either a defect in initiating the early gene expression program or trafficking of the cAMP receptor to the membrane (Fig. S2 A and B). The myoG mutants are specifically impaired in their response to cAMP stimulation because of a defect in early signal transduction events from the G protein-coupled receptor cAR1 to the actin cytoskeleton. Chemotaxis to both folate and cAMP relies on a common Gβγ complex, but each requires distinct Gα subunits for downstream signaling (30–32). The observation that myoG nulls move toward folate (Fig. 1) suggests that MyoG does not have a role in signaling from Gβγ to the folic acid chemotaxis machinery. The mutants exhibit a weak increase in cortical actin polymerization and fail to localize and activate PI3K in response to cAMP stimulation (Fig. 4). These data indicate that the myoG mutant defect lies upstream of both of these crucial events in response to stimulation by cAMP and implicates MyoG in an early chemotactic signaling pathway in Dictyostelium.
The two main features of chemotaxis, cellular polarization and directed movement toward the source of cAMP, are absent in the myoG null mutant. Both behaviors depend on highly localized signaling from the cAR1 receptor-coupled heterotrimeric G-protein complex to the actin cytoskeleton. Dissociation of the G-protein complex from the activated receptor stimulates guanine nucleotide exchange factors (GEF)-mediated activation of small GTPases, which in turn promotes localized actin polymerization at the region of the membrane closet to attractant and M2 filament assembly at the rear of the cell (21). The suppressed initial burst of actin polymerization following stimulation and absence of PI3K localization to the membrane closest to the cAMP source observed in the myoG mutant suggests that upstream signaling events are severely compromised in the absence of MyoG. RasG lies upstream of PI3K and it is required for full activation of this key kinase (28). However, the rasG single mutant only exhibits a mild defect in chemotaxis and cell polarization. The small G protein RacB mediates changes in actin polymerization in response to cAMP potentially by activating (directly or indirectly) the Arp2/3 regulator WASP (33). The racB mutants exhibit the same actin polymerization profile as that observed in myoG mutants, a dampened early peak of actin polymerization and a missing second peak. Similar to what was observed for the rasG mutant, aggregation of the racB mutants is only delayed and they do proceed to form aggregates and fruiting bodies. Dictyostelium expresses a number of regulators of small G protein activity, and RasGEFR and RacGEF1 are both implicated in chemotactic signaling, but deletion of either alone does not completely abrogate chemotactic responses (34). The phenotypes of the different signaling mutants suggest that a combined loss of activity from RacGEFR and RacGEF1 pathways would be cumulative, resulting in the complete inability of cells to polarize and chemotax. MyoG may modulate the activity of several key small G proteins by controlling the activation or localization of specific GEFs (and also GTPase activating proteins) at the membrane, possibly by serving as a central component of a signaling hub that relays heterotrimeric G protein activation to GEFs and key downstream small GTPases to stimulate localized actin polymerization and PI3K activation.
The social amoebae and mammalian lineages appear to have arisen from a common ancestor, accounting for the fundamental conservation in cellular function and signaling pathways between Dictyostelium and vertebrates (35). A true MyoG homolog is not present in higher eukaryotes; however, the striking conservation of chemotactic signaling pathways between amoeba and vertebrate cells suggests that a functional homolog may exist. In fact, MyTH/FERM proteins have recently been found to have roles in chemotactic signaling in both mice and worms. M10 contributes to axonal pathfinding in mouse cortical neurons (36). Neurons lacking M10 are completely deficient in neurite outgrowth normally observed in response to the guidance cue netrin-1. M10 is also necessary for BMP-induced directed migration of mouse intraembryonic endothelial cells (37), where it is not only required for filopod formation in response to BMP, but it also has a role in mediating the intracellular response to BMP. The PH/MyTH/FERM protein MAX-1 mediates repulsive cues during axonal guidance in Caenorhabditis elegans, where it potentially participates in signaling from the UNC-5 netrin receptor (38). Together, these data suggest that MyTH/FERM myosins contribute to different types of chemotactic signaling and that MyTH/FERM proteins may have general and widespread roles in responding to guidance cues and the generation of cell polarity.
Materials and Methods
Cell Growth and Development.
Dictyostelium strains were maintained using standard methods (39). Cells were grown on tissue culture plates in HL5 growth medium supplemented with 10,000 U/mL penicillin G (Fisher Scientific) and 10 μg/mL streptomycin sulfate (Sigma Chemical Co.). The myoG null mutant (HTD36-2) and the NHR control strains were maintained under constant selection in HL5 supplemented with 10 μg/mL Blasticidin S (ICN Biomedicals). Transformants expressing coronin-GFP (40), CRAC-GFP (41), N-PI3K1-GFP (28), or cAR1-GFP (42) in the wild-type Ax2, NHR control, or myoG null mutant backgrounds were maintained in HL5 supplemented with 10 μg/mL G418 (Fisher Scientific). Aggregation competent cells were obtained by washing and resuspending log phase cells in MMC (20 mM Mes, 2 mM MgSO4, 0.2 mM CaCl2) or LPS (20 mM KCl, 0.24 mM MgCl2, 40 mM Na2HPO4, pH 6.4) to a final density of 1 to 2 × 107 cells/mL. The cells were shaken at 150 to 200 rpm for 1 h, and then 50 to 100 nM cAMP pulses were administered every 6 min for 4 to 7 h (41).
Phenotypic Analysis.
Chemoattractant-stimulated changes in actin polymerization were assayed as described (43). Fluorescence was measured with a Wallac Victor plate reader (excitation 530 nm, emission 590 nm) (PerkinElmer). For F-actin visualization, vegetative cells adhered to coverslips were formaldehyde-fixed and stained with 1:500 Alexa 488-phalloidin (Invitrogen) as described (44). Confocal images were obtained with a Nikon TE200 microscope equipped with a Plan-Apo 60×, 1.40 NA oil-immersion objective. cAR1-GFP expressing vegetative cells were allowed to adhere to coverslips, washed with MMC, and photographed within 30 min. Mixing experiments were performed using aggregation competent cultures consisting of 85% Ax2 cells and 15% of either NHR cells or myoG mutant cells expressing coronin-GFP. The cell mixture was allowed to adhere to coverslips and fluorescent and differential interferenced contrast images taken once streaming had begun. Aggregation competent CRAC-GFP- and N-PI3K1-GFP-expressing cells were adhered to the bridge of a Zigmond chamber (Neuro Probe) and exposed to a cAMP gradient as described below. Images were obtained with a Zeiss Axiovert microscope and 63× Plan-APO 1.4 NA oil-immersion lens. A small-drop folic acid chemotaxis assay on vegetative cells was performed as described (18). Chemotaxis to cAMP was assayed by washing aggregation-competent cells twice with LPS followed by dilution to 5 × 105 cells /mL for chemotaxis analysis and 2.5 × 105 cells /mL for perfusion analysis. A Sykes-Moore perfusion chamber (Bellco Glass) was used to analyze cell behavior in the absence of chemoattractant, as described (45). Cells adhered to a 25-mm glass coverslip inside the chamber were perfused with LPS at a rate that turned over one chamber volume-equivalent every 15 s. Behavior of cells in a spatial gradient of cAMP was analyzed in a Plexiglass chamber designed after that of Zigmond (46, 47). LPS alone was added to one trough; LPS containing 1 μM cAMP was added to the other trough and cells were recorded after 10 to 15 min in the chamber. Two-dimensional Dynamic Image Analysis System software was used for all analysis of cell behavior (22).
Supplementary Material
Acknowledgments
The authors thank William Manzel, Kathleen Mahan, Zachary Ryan, Casey Dorr, and Dr. Heather Francis for assisting during the early stages of this project, Spencer Kuhl for technical assistance with DIAS, and Drs. Peter Devreotes (Johns Hopkins) and Rick Firtel (University of California San Diego) for generously providing some of the reporter plasmids used in this study. This work was supported in part by National Institutes of Health Training Grant in Developmental Biology T32-HD007480 (to L.M.B.), a Doctoral Dissertation Fellowship from the Graduate School of the University of Minnesota (L.M.B.), and by grants from the National Institutes of Health (to D.R.S.), National Science Foundation Grant MCB-0424704 (to M.A.T.), and American Heart Association Grant 00004962 (to M.A.T.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909796107/DCSupplemental.
References
- 1.Affolter M, Weijer CJ. Signaling to cytoskeletal dynamics during chemotaxis. Dev Cell. 2005;9:19–34. doi: 10.1016/j.devcel.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. 2008;9:960–969. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 3.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 4.Narasimhulu SB, Reddy AS. Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell. 1998;10:957–965. doi: 10.1105/tpc.10.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 6.Chishti AH, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 7.El-Amraoui A, Bahloul A, Petit C. In: Myosin VII. Myosins: A Superfamily of Molecular Motors. Coluccio LM, editor. Dordrecht, The Netherlands: Springer; 2008. pp. 353–373. [Google Scholar]
- 8.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 9.Liu R, et al. Sisyphus, the Drosophila myosin XV homolog, traffics within filopodia transporting key sensory and adhesion cargos. Development. 2008;135:53–63. [Google Scholar]
- 10.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Probst FJ, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- 12.Tuxworth RI, et al. A role for myosin VII in dynamic cell adhesion. Curr Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 13.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol Cell Biol. 2008;28:1702–1712. doi: 10.1128/MCB.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerber ML, et al. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokuo H, Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem Biophys Res Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 16.Belyantseva IA, et al. Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7:148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- 17.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segall JE, Fisher PR, Gerisch G. Selection of chemotaxis mutants of Dictyostelium discoideum. J Cell Biol. 1987;104:151–161. doi: 10.1083/jcb.104.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chisholm RL, Firtel RA. Insights into morphogenesis from a simple developmental system. Nat Rev Mol Cell Biol. 2004;5:531–541. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- 20.Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- 21.Franca-Koh J, Kamimura Y, Devreotes P. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr Opin Genet Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Wessels D, Kuhl S, Soll DR. Application of 2D and 3D DIAS to motion analysis of live cells in transmission and confocal microscopy imaging. Methods Mol Biol. 2006;346:261–279. doi: 10.1385/1-59745-144-4:261. [DOI] [PubMed] [Google Scholar]
- 23.Hall AL, Schlein A, Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J Cell Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- 24.McRobbie SJ, Newell PC. Changes in actin associated with the cytoskeleton following chemotactic stimulation of dictyostelium discoideum. Biochem Biophys Res Commun. 1983;115:351–359. doi: 10.1016/0006-291x(83)91011-2. [DOI] [PubMed] [Google Scholar]
- 25.Comer FI, Lippincott CK, Masbad JJ, Parent CA. The PI3K-mediated activation of CRAC independently regulates adenylyl cyclase activation and chemotaxis. Curr Biol. 2005;15:134–139. doi: 10.1016/j.cub.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 27.Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki AT, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadwiger JA, Lee S, Firtel RA. The G alpha subunit G alpha 4 couples to pterin receptors and identifies a signaling pathway that is essential for multicellular development in Dictyostelium. Proc Natl Acad Sci USA. 1994;91:10566–10570. doi: 10.1073/pnas.91.22.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai A, Hadwiger JA, Pupillo M, Firtel RA. Molecular genetic analysis of two G alpha protein subunits in Dictyostelium. J Biol Chem. 1991;266:1220–1228. [PubMed] [Google Scholar]
- 32.Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park KC, et al. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004;23:4177–4189. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortholt A, van Haastert PJ. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 2008;20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Schaap P, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu XJ, et al. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9:184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
- 37.Pi X, et al. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179:1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Cheng HJ, Tessier-Lavigne M, Jin Y. MAX-1, a novel PH/MyTH4/FERM domain cytoplasmic protein implicated in netrin-mediated axon repulsion. Neuron. 2002;34:563–576. doi: 10.1016/s0896-6273(02)00672-4. [DOI] [PubMed] [Google Scholar]
- 39.Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 40.Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 41.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Z, Zhang N, Murphy DB, Devreotes PN. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J Cell Biol. 1997;139:365–374. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JY, et al. Phosphorylation of chemoattractant receptors is not essential for chemotaxis or termination of G-protein-mediated responses. J Biol Chem. 1997;272:27313–27318. doi: 10.1074/jbc.272.43.27313. [DOI] [PubMed] [Google Scholar]
- 44.Peterson MD, Novak KD, Reedy MC, Ruman JI, Titus MA. Molecular genetic analysis of myoC, a Dictyostelium myosin I. J Cell Sci. 1995;108:1093–1103. doi: 10.1242/jcs.108.3.1093. [DOI] [PubMed] [Google Scholar]
- 45.Varnum B, Edwards KB, Soll DR. The developmental regulation of single-cell motility in Dictyostelium discoideum. Dev Biol. 1986;113:218–227. doi: 10.1016/0012-1606(86)90124-7. [DOI] [PubMed] [Google Scholar]
- 46.Varnum B, Soll DR. Effects of cAMP on single cell motility in Dictyostelium. J Cell Biol. 1984;99:1151–1155. doi: 10.1083/jcb.99.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.