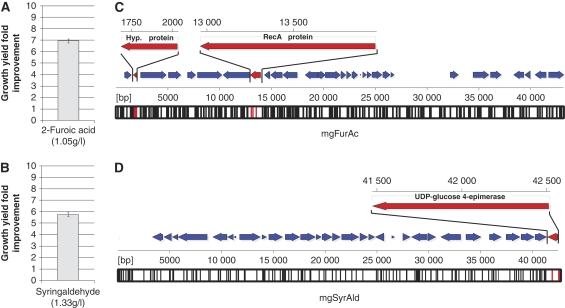
Figure 2.
Sequence annotation and functional analysis of selected genetic elements improving biomass inhibitor tolerance in E. coli. (A, B) Improvements in inhibitor tolerance toward 2-furoic acid and syringaldehyde because of metagenomic inserts. Inhibitor concentrations resulting in 90% reductions in growth yield were determined for wild-type E. coli as 1.05 g/l for 2-furoic acid and 1.33 g/l for syringaldehyde. Improvements in E. coli growth yield at these concentrations because of metagenomic inserts were 6.9-fold for 2-furoic acid and 5.7-fold for syringaldehyde, showed here as the mean (and standard deviation) of triplicate readings after 24 h of growth. (C, D) The metagenomic inserts conferring tolerance to 2-furoic acid (mgFurAc) and syringaldehyde (mgSyrAld) were sequenced at 3 × coverage and annotated (Supplementary Tables II and III). Annotated genes for (C) mgFurAc and (D) mgSyrAld are shown as filled arrows, with the orientation denoting the relative direction of transcription based on an arbitrary sense strand. Transposon mutagenesis, followed by reselection of the tolerance phenotypes, was used to identify functional genetic elements in mgFurAc and mgSyrAld that contribute to the selected phenotypes (genes colored red and labeled) (Supplementary information). Vertical bars along the bottom of each sequence–position axis denote positions of transposon insertion in the loss-of-function study (black denotes no effect, red denotes loss-of-function).