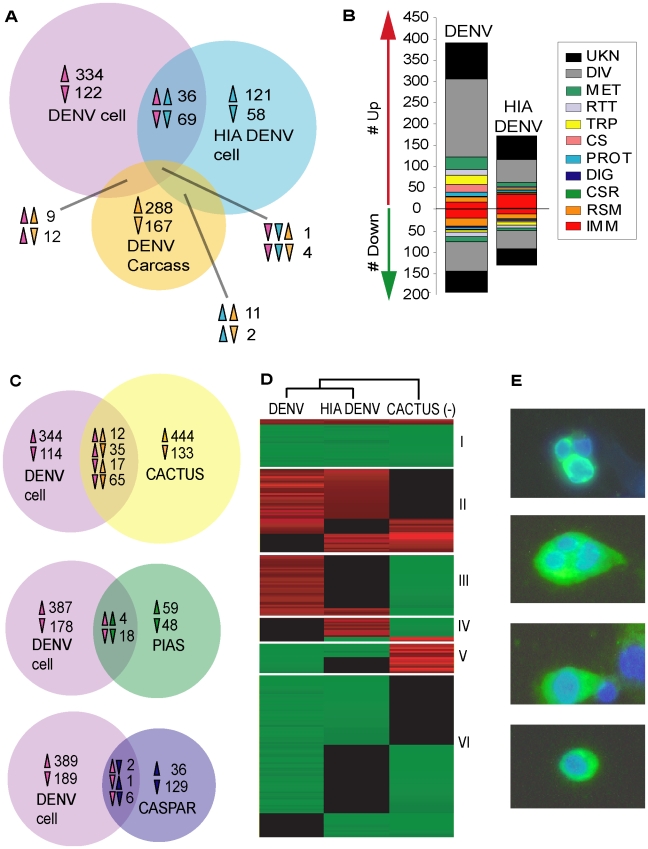
Figure 1. Transcriptional regulation of genes in the Aag2 cell line in response to live dengue virus (DENV) and heat-inactivated dengue virus (HIA DENV) infection.
A. Venn diagram showing the numbers of unique and commonly regulated genes in DENV- and HIA DENV-infected cells and DENV-infected mosquito carcass. Arrows indicate the direction of gene regulation. B. Functional classification of significantly regulated genes in DENV and HIA DENV infection; arrows indicate the direction of gene regulation. Functional group abbreviations are as follows: UNK, unknown functions; DIV, diverse functions; MET, metabolism; RTT, replication, transcription, and translation; TRP, transport; CS, cytoskeletal and structural; PROT, proteolysis; DIG, blood and sugar food digestive; CSR, chemosensory reception; RSM, redox, stress and mitochondrion; IMM, immunity. C. Comparative analysis of the DENV infection-responsive cell line transcriptome and the Toll-, IMD-, and JAK-STAT pathway-regulated mosquito transcriptomes. Venn diagrams show the numbers of unique and commonly regulated genes in DENV-infected Aag2 cells and Cactus-, Caspar-, and PIAS-silenced A. aegypti mosquitoes. Arrows indicate the direction of gene regulation. D. Cluster analysis of 238 genes that were regulated in at least two of three treatments: DENV infection in the cell line, HIA DENV infection in the cell line, Cactus silencing in A. aegypti mosquitoes. All genes presented in this cluster analysis are listed in Table S6. E. Detection of DENV in Aag2 cells by indirect immunofluorescence assay using mouse hyperimmune ascitic fluid specific for DENV2 (CDC) and an AlexaFluor488-conjugated goat anti-mouse secondary antibody. Blue: DAPI, Green: FITC.