Abstract
Unaccustomed eccentric exercise damages skeletal muscle tissue, activating mechanisms of recovery and remodeling that may be influenced by the female sex hormone 17β-estradiol (E2). Using high density oligonucleotide based microarrays, we screened for differences in mRNA expression caused by E2 and eccentric exercise. After random assignment to 8 days of either placebo (CON) or E2 (EXP), eighteen men performed 150 single-leg eccentric contractions. Muscle biopsies were collected at baseline (BL), following supplementation (PS), +3 hours (3H) and +48 hours (48H) after exercise. Serum E2 concentrations increased significantly with supplementation (P<0.001) but did not affect microarray results. Exercise led to early transcriptional changes in striated muscle activator of Rho signaling (STARS), Rho family GTPase 3 (RND3), mitogen activated protein kinase (MAPK) regulation and the downstream transcription factor FOS. Targeted RT-PCR analysis identified concurrent induction of negative regulators of calcineurin signaling RCAN (P<0.001) and HMOX1 (P = 0.009). Protein contents were elevated for RND3 at 3H (P = 0.02) and FOS at 48H (P<0.05). These findings indicate that early RhoA and NFAT signaling and regulation are altered following exercise for muscle remodeling and repair, but are not affected by E2.
Introduction
Myofibres have the capacity to be remodeled to best meet their functional and metabolic demands. Changes in physical activity can initiate a remodeling process toward increased (hypertrophy) or decreased (atrophy) muscle mass [1], [2], [3]. Through protein signaling pathways, integration of chemical, mechanical and bioenergetic signals change the genetic expression patterns for cell size, function and metabolic processes [4]. Physical activities that incorporate unaccustomed eccentric contractions are typically associated with high levels of muscle damage, inflammation and delayed onset muscle soreness (DOMS) [5], [6], [7]. Eccentric contractions, identified by a lengthening while under tension, create an insult to myofibres which may be characterized by: damage to the sarcoplasmic reticulum [5], t-tubules [8] and structural proteins [6], [9], [10], the presence of muscle protein in blood [11], [12], Z-line streaming [13], [14], soreness and fatigue [7], [8].
Murine and rodent research has often indicated an attenuation of exercise induced membrane damage [15], [16], [17], [18], [19], structural proteins [20], and inflammation [21], [22], [23], [24] along with enhanced satellite cell activation [21], [25], [26] with exposure to the sex hormone 17β-estradiol (E2). This reduced muscle damage and improved recovery may result from potential antioxidant, membrane stabilizing, or gene regulation properties of E2 [27], [28], [29], [30], [31]. As the most abundant estrogen, E2 exerts estrogenic properties that affect the differentiation, growth and function of reproductive, skeletal, neural and muscular tissues [31], [32]. However, human studies do not consistently support the effectiveness of E2 to attenuate exercise-induced muscle damage, mostly reporting similar values for CK efflux, inflammation and loss of muscle function when comparing men and women [6], [33].
Recent microarray analyses have uncovered novel transcriptional programs that coordinate the regeneration and repair of damaged muscle following eccentric exercise [10], [34], [35]. These studies have indentified clusters of genes representing important mechanisms for recovery and adaptation that include the regulation of inflammation [10], [35], growth [10], [35], stress response proteins [10], [34] and membrane biosynthesis [34]. Similar analyses have also described sex differences in the expression levels of genes involved in metabolism and growth inhibition that may result from variations in body composition and hormone content [36], [37], [38]. Specifically, women express a higher abundance of mRNA for several genes involved in fat metabolism that include trifunctional protein β and lipoprotein lipase [36], and greater expression of the negative regulators of the anabolism growth factor receptor-bound 10 and activin A receptor IIB [37].
The effect of E2 administration on the transcriptome expression profile of skeletal muscle following a single bout of intense eccentric exercise has not yet been evaluated. In this study we used microarray analysis to identify how global mRNA abundance is altered by E2 supplementation in men after 150 eccentric contractions. We hypothesized that the anti-oxidant and membrane stabilizing properties of E2 would attenuate the amount of muscle damage experienced, thereby modifying the expression of mRNA species involved in membrane homeostasis, growth and stress management. Furthermore, we hypothesized that the use of gene array analysis would allow us to detect novel genes relevant to the hypertrophic growth signaling stimulated by intense exercise.
Materials and Methods
Ethics statement
All participants were given an information sheet describing all of the testing procedures before providing written consent to participate. The study conformed to the standards outlined in the Declaration of Helsinki and was given approval by the Research Ethics Board of McMaster University (05–438).
Subjects and anthropometrics
Eighteen young healthy men volunteered as participants in this study. All subjects were pre-screened to ensure that they were healthy, fit and had not regularly participated in resistance exercise in the preceding 6 months. Body composition was measured using dual energy x-ray absorptiometry (DEXA) scans (GE Lunar Prodigy, Fairfield, CT). Thigh muscle cross-sectional area was calculated using anthropomorphic measurements of mid-thigh circumference and skinfold thickness [39] to control for potential differences in total work completed. The subject demographics were (mean ± SD): age, 21±2 y; height, 181±5 cm; weight, 76.9±12.8 kg.
Supplementation protocol
Subjects were assigned in a randomized, double-blind manner to either a control (CON, N = 9) or experimental (EXP, N = 9) group. CON subjects consumed 400 mg glucose polymer (Polycose; Abbott Laboratories, Ross Division, St. Laurent, Quebec, Canada) for 10 days. EXP subjects consumed ∼300 mg glucose with 1 mg E2 (Estrace; Shire BioChem, Inc., St. Laurent, Quebec, Canada) for 2 days followed by 2 mg E2 for 8 days. We have previously used this protocol to increase serum E2 concentrations to levels seen during the luteal phase of the menstrual cycle [40]. Glucose and E2 tablets were concealed in gelatin capsules. On the morning of the ninth day, subjects reported to the laboratory and performed the exercise protocol. Supplementation continued until the day of the final biopsy and blood collection to maintain serum E2 concentrations throughout the collection protocol. Subjects in both groups were instructed to take one pill at the same time each day and return any unused pills. All subjects reported 100% compliance.
Exercise protocol and tissue collection
Muscle damage was induced with a previously developed eccentric exercise protocol [41]. Approximately 2 weeks before the exercise protocol, subjects were given a familiarization session with a Biodex isokinetic dynamometer (System 3, Biodex Medical Systems Inc., Ronkonkoma, NY). On the testing day, following a short warm-up (10 min of light cycling), subjects were seated in the dynamometer with their right leg strapped to a lever arm. The lever arm was programmed to extend their leg to 150° of flexion (where 180° is full extension) at a moderate speed (30°/s), then flex their leg to 90° of flexion at a faster speed (120°/s). Subjects did not have to contract maximally during the extension phase. During the flexion phase, subjects were instructed to attempt to maximally resist flexion of the knee (i.e. voluntary ‘maximal’ contraction) against the descending lever arm throughout the entire range of motion. The complete test consisted of 15 sets of 10 repetitions, each set separated by 1 minute of rest.
Prior to each tissue collection, subjects abstained from any other form of physical exertion (within 72 h), avoided alcohol (within 48 h), ate their habitual diet (within 48 h), and abstained from caffeine (within 12 h). Each subject consumed a 350 Kcal defined formula diet (57% carbohydrates, 15% protein and 28% fat) two hours before each muscle biopsy and did not eat again until after the final biopsy of each session was taken. These nutritional and activity controls were taken to ensure that the muscle damage would be the only variable to differentially affect the outcomes between biopsies [42].
Muscle biopsies were taken from the vastus lateralis of the control (left) leg during the familiarization session (baseline, BL) and after 8 days of supplementation (post supplementation, PS) and the exercised (right) leg 3 hours (3H) and 48 hours (48H) after exercise, in anatomically distinct sites approximately 6 cm apart [43]. The post exercise collection times were chosen because they represent two distinct phases of recovery from muscle damage [6]. The muscle biopsies were quickly dissected of fat and connective tissue, sectioned into RNase-free cryovials (∼30 mg/piece), flash frozen with liquid nitrogen and stored at −86°C until analysis. Blood samples were drawn from the antecubital vein into heparinized tubes at the same collection times, placed on ice, centrifuged at 1750 g at 4°C for 10 min and stored at −20°C future analyses.
Blood hormone and enzyme concentrations
Serum E2 (Fertigenix-E2-EASIA, Biosource Europe S.A, Nivelles, Belgium) and testosterone (Fertigenix-TESTO-EASIA, Biosource Europe S.A, Nivelles, Belgium) concentrations were measured by enzyme amplified-sensitivity immunosorbent assays (EASIA) according to manufacturer's specifications using BL and PS blood collections. Serum lactate dehydrogenase (LDH) activities were measured in BL, 3H and 48H blood collections with a colourimetric LDH quantification assay (K726-500, Biovision Research Products, Mountain View, CA) according to manufacturer's specifications. All hormone and enzyme measurements were done in duplicate.
RNA extraction
The total RNA was extracted from the frozen skeletal muscle biopsy as described previously in detail by our group [43]. Briefly, ∼30 mg of skeletal muscle was homogenized on ice in 2 mL of Trizol Reagent (Life Technologies, Cat. No. 15596, Gaithersburg, MD). The homogenate was incubated for 10 min at room temperature, followed by phase separation using 200 µL of chloroform and precipitation of the total RNA from the aqueous phase using 500 µL of isopropyl alcohol. The RNA pellet was then washed three times in 75% ethanol and re-suspended in 15 µL DEPC-treated water, aliquoted, and stored at −86°C. The concentration and purity of the RNA was determined using a UV spectrophotometer (Shimadzu UV-1201; Mandel Scientific, Guelph, Ontario) at the absorbance of 260/280 nm. Measurements were done in duplicate and had an average coefficient of variation (CV) of <10%. The average purity (OD260/OD280) of the samples was 1.7 before DNase treatment. RNA integrity was assessed in a randomly chosen subset of samples using agarose gel electrophoresis, and the OD ratio of 28S to 18S rRNA was consistently greater than 1 for each sample.
DNase treatment
Prior to microarray chipping and real time quantitative RT-PCR analysis, the isolated RNA samples were treated with DNA-free™ recombinant DNase I (Ambion Inc, Austin, TX) according to the manufacturer's instructions to remove any potential genomic DNA contamination.
Microarray analysis
The resulting total RNA samples were further assessed for integrity prior to chipping using a Nanodrop Spectrophotometer and the Agilent Bioanalyzer Nano Chip System. Samples which passed this initial quality control assurance step were then amplified one round, using an Illumina TotalPrep Kit (Ambion) to generate cDNA then cRNA according to the manufacturer's instructions. This was again assessed for quality by using the Nanodrop and Bioanalyzer as described above. Labeled cRNA samples that passed this second round of quality control were then hybridized to Human Ref-8 BeadChips (Illumina) according to the manufacturer's instructions (approximately 23,000 genes), using equipment specified by the manufacturer (Illumina). Briefly, 850 ng biotin-labeled cRNA in 11.3 µl nuclease-free water was adjusted to 34 µl through the addition of 22.7 µl of 5∶3 HybE1 buffer/formamide. The sample was heated at 65°C for 5 min, allowed to cool to room temperature, and then immediately added to a single array of an 8-array Human Ref-8 BeadChip. Once all 8 samples were added to each BeadChip, it was sealed in a Hyb Cartridge and incubated for 16 h at 55°C with rotation in an Illumina hybridization oven (rotation setting 5). Following overnight hybridization, BeadChips were moved to a slide rack and serially washed using gentle rotation in glass staining dishes filled with a) 250 ml Illumina Wash Buffer×5 min, b) 250 ml 100% ethanol×10 min, c) 250 ml Illumina Wash Buffer×2 min. BeadChips were then blocked for 10 min in 4 ml Block E1 buffer (Illumina), followed by staining for 10 min in 1 µg/ml Streptavidin-Cy3 conjugate (GE Healthcare) in Block E1 buffer. Stained BeadChips were finally washed using gentle rotation in a glass staining dish filled with 250 ml Illumina Wash Buffer×5 min. BeadChips were dried by centrifugation at 280 g for 4 min and stored in a light-tight box until reading.
Array reading
Processed arrays were read using a BeadStation array reader (Illumina) according to the manufacturer's instructions.
Gene ontology analysis
In the lists of genes that were significantly differentially expressed with exercise in our study, we carried out gene ontology (GO) analysis to determine the relative enrichment of genes with common or related functionalities to gain insight into biological processes mediated by E2 or exercise. This was carried out using the web interface driven GoMiner tool using an FDR of 5% [44]. Genes were also referenced to their biological functions and canonical pathways with Ingenuity Pathway Analysis (IPA) software. This software identifies functions and pathways most significant to the data set in two ways: the number of differentially expressed genes included in a pathway or function and calculation of a p-value using a Fisher's Exact Test to determine the probability of the association of the data set.
Real-time RT-PCR analysis
Changes in gene expression relative to baseline values were measured using real-time reverse transcription-polymerase chain reaction (RT-PCR). Regulator of calcineurin 1 (RCAN1) and capping protein (actin filament) muscle Z-line, alpha 1 (CAPZA1) were selected for analysis because of their roles in growth and sarcomerogenesis [34]. Hemeoxygenase 1 (HMOX1) was chosen for analysis because of its role in stress management [45]. The selected housekeeping gene was β2-microglobulin. Its constant expression following eccentric exercise has been shown in previous work [43], and was confirmed for the current study. The efficiencies of all primers were tested and determined to be greater than 98%. The primer and probe sequences for these genes can be found in Table 1.
Table 1. Primer and probe sequences for calcineurin regulation, actin dynamics and housekeeping genes.
| Gene | Left Primer | Right Primer | Probe |
| RCAN1 | gacaaggacatcacctttcagt | tcatttcctttcccagaaactc | caaacgagtcagaataacttcagcaaccc |
| HMOX1 | tctccgatgggtccttacac | cctgcattcacatggcataa | ctaagccaactgtcgccaccagaaa |
| CAPZA1 | ccttccaagcacttctggtact | gagggagaaggatgaatgtgt | atccaccaacacctaaagaggctatgc |
| β2M | ggctatccagcgtactccaa | gatgaaacccagacacatagca | tcaggtttactcaacgtcatccagcagag |
RT-PCR was completed using a TaqMan® real-time method. The primers and a probe to each target gene were designed based on the cDNA sequence in GenBank (http://www.ncbi.nlm.nih.gov/sites/entrez/?db=gene) with primer 3 designer (http://frodo.wi.mit.edu/primer3-0.4.0/input.htm). All target gene probes were labeled with FAM at their 5′ ends and BHQ-1 at their 3′ ends. Duplex RT-PCR was performed on an iCycler real-time PCR system (Bio-Rad Laboratories, Hercules, CA) in the One-step TaqMan® RT-PCR Master Mix Reagents (Roche, Branchburg, New Jersey) according to the manufacture's instruction with target gene primers, target probe, housekeeping gene primers and housekeeping gene probe in the same reaction [46]. Determination of significant gene expression change was done as previously described [46]. The genes of interest were normalized to the housekeeping gene, β2-microglobulin by following the standard method. Briefly, CT values of the housekeeping gene were subtracted from the CT values of the gene of interest giving a δCT. This is equivalent to the log2 difference between endogenous control and target gene [47]. Values were then normalized to baseline, δδCT. All samples were run in triplicate, fluorescence emission was detected using FAM and Tamra filters, and CT was automatically calculated.
Western blotting
Muscle biopsy samples were homogenized and prepared for polyacrylamide gel electrophoresis using methods previously described [48]. Briefly, frozen skeletal muscle tissue samples (25–35 mg) were hand homogenized in 25 µl of phosphate buffer (50 mM Kpi, 5 mM EDTA, 0.5 mM DTT, 1.15%KCl (w/v)) per milligram of tissue. A protease inhibitor cocktail (Sigma, St. Louis, Missouri) was added to the phosphate buffer immediately prior to use at a ratio of 1∶1,000. Samples were centrifuged at 600 g for 10 min at 4°C and the supernatant aliquoted for analyses. Protein concentrations of each sample were determined using the method described by Lowry et al [49].
Samples were loaded on 10% SDS-polyacrylamide gels and transferred to a PVDF membrane. Membranes were blocked with 5% BSA (wt/vol) in Tris-buffered saline with 0.1% Tween (vol/vol) (TBST) and incubated in primary antibody: RND3 (Abcam, Cambridge, MA; ab50316, 1∶1000); FOS (Abcam; ab16902, 1∶1000); total p38MAPK (Cell Signaling Technology, Danvers, MA; no. 9212, 1∶1000); p38MAPK (Thr180/Tyr182) (Cell Signaling Technology; no. 9215, 1∶1000); total GSK-3β (Cell Signaling Technology; no. 9315, 1∶1000); GSK-3β (Ser 9) (Cell Signaling Technology; no. 9323, 1∶1000), β-actin (BD Biosciences, Mississauga, ON; no. 612657, 1∶10000). After washing in TBST, membranes were incubated in either HRP-linked anti-rabbit or anti-mouse IgG secondary antibody (Amersham Biosciences, Piscataway, NJ; no. NA934V, 1∶6000), washed with TBST and developed using ECL (Amersham Biosciences; model no. RPN2106). Membranes were exposed to x-ray film (Biomax XAR; Kodak, Rochester, New York) which were then scanned with a Dell 920 scanner at 300 DPI and saved in TIF file format. Using Image J v1.40 g software (National Institutes of Health, Bethesda, Maryland), background noise was removed and bands in the region of interest were selected for analysis. Individual profile plots were generated and area under the curve measured in arbitrary units (AU).
Statistical and bioinformatics analysis
Student's unpaired t-tests were used to determine differences in subject characteristics and total work. A 2-way repeated measures ANOVA (supplementation protocol × time) was used to assess differences in LDH, E2 concentration, testosterone concentration, protein levels and the linear 2−δδCT data set for gene expression measured with RT-PCR using computerized software (Statistica, Statsoft). When statistical significance was achieved, Tukey's honestly significance difference post-hoc test was used to determine the significance among the means. STATISTICA for Windows 5.0 (Statsoft, Tulsa OK) was used to perform t-tests and ANOVAs. The threshold for significance was set at P≤0.05. Data are presented as mean ± SEM unless otherwise indicated.
Gene array data analyses were done comparing baseline, post supplementation, 3 and 48 hours post exercise using simple paired t-test on log2 expression ratios. Genes were ranked by p-value and the inference reported following adjustment for multiple testing using the FDR and the Benjamini and Hochberg method. Among those genes with an adjusted q-value (based on FDR) of <0.05, we used hierarchical clustering (based on the HOPACH algorithm) to find groups of genes with similar profiles across the subjects.
Results
Subject and work characteristics
CON and EXP groups were not different in age, weight, height, body fat percentage, average thigh cross-sectional area or total work completed (Table 2). All subjects completed the required 150 eccentric contractions.
Table 2. Subject and eccentric exercise trial characteristics.
| CON | EXP | |
| No. of subjects | 9 | 9 |
| Age, yr | 21.1±0.8 | 20.9±0.9 |
| Height, cm | 181.6±2.0 | 180.9±1.4 |
| Weight, kg | 73.4±3.8 | 80.4±4.6 |
| Body fat, % | 14.7±1.7 | 20.1±2.4 |
| Quadriceps CSA, cm2 | 74.4±3.3 | 78.5±4.1 |
| Work, kJ | 24.9±2.9 | 25.1±1.3 |
Values are means ± SEM.
E2 and testosterone concentration were affected by supplementation with E2
Following 8 days of supplementation serum E2 concentration increased by 146% (P<0.001) and testosterone concentration was reduced by 26% (P = 0.01) in the EXP group (Table 3). Both hormone concentrations remained unchanged in the CON group.
Table 3. Serum hormone concentrations after 8d of supplementation with either placebo (CON) or E2 (EXP).
| Estradiol (pg/ml) | BL | PS |
| CON | 36.4±2.5 | 38.4±2.8 |
| EXP | 38.8±4.0 | 95.4±11.7*** |
Values are mean ± SEM.
P<0.01.
P<0.001.
N = 9/group.
Eccentric exercise induced muscle damage
The appearance of the muscle protein LDH in serum is an indirect indicator of muscle membrane damage. LDH activity was elevated 13.8% (P<0.05) 48 hours after exercise (Table 4). The EXP values did not differ from the CON values at any time.
Table 4. Serum lactate dehydrogenase activity following 150 eccentric contractions in CON and EXP groups.
| Baseline | 3 hours | 48 hours* | |
| CON (U/L) | 152.3±8.6 | 166.1±6.8 | 166.3±10.9 |
| EXP (U/L) | 140.7±9.7 | 158.1±9.2 | 165.4±16.3 |
Values are mean ± SEM.
*P<0.05 main effect for group compared to baseline.
CON (N = 7), EXP (N = 8).
Microarray data identifies altered mRNA expression during recovery from eccentric exercise that is not affected by E2
Eccentric exercise significantly increased the early mRNA expression of 310 genes at 3H. DNA microarray analyses did not identify differential mRNA expression of any gene as a result of E2 at any time (we could not reject the global null that E2 was independent of mRNA expression of all genes represented on the chip). For this reason, microarray data at each timepoint was collapsed between groups, increasing the sample size to 18 subjects. Mean fold change for genes with ratios greater than 1 at 3H was 2.3±0.1. In addition, 301 genes had ratios less than 1 at 3H with a mean fold change of 0.8±0.01. By 48H, all genes had expression values that were not different from baseline. The complete data set is freely available at GEO (accession no. GSE19062).
Of the genes differentially expressed at 3H, we identified 25 that participate in two signaling cascades for muscle growth and adaptation: ras homologue gene family, member A (RHOA) and nuclear factor of activated T-cells (NFAT) (Table 5). Other regulators of muscle growth and remodeling not directly involved in RHOA or NFAT signaling that were also highly induced included: ATF3, MYC, XIRP1, HBEGF and DNAJB4 (Table 5).
Table 5. Fold change of gene expression after 3 hours of recovery from eccentric exercise using DNA microarray analysis (n = 18).
| Categories and Gene Names | Accession Number | Fold change at 3H | Potential relevant function |
| Calcineurin regulation | |||
| regulator of calcineurin 1 (RCAN1), transcript variant 3 | NM_203418.1 | 5.6 | Calcinuerin regulation |
| regulator of calcineurin 1 (RCAN1), transcript variant 2 | NM_203417.1 | 3.9 | Calcinuerin regulation |
| NFAT | |||
| nuclear factor of activated T-cells, (NFATC1), transcript variant 1 | NM_172390.1 | 1.3 | calcineurin transcription/regulation |
| glycogen synthase kinase 3 beta (GSK3B) | NM_002093.2 | 1.3 | calcineurin transcription/regulation |
| MAPK regulation | |||
| Activation | |||
| mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2), transcript variant 2 | NM_032960.2 | 1.2 | MAPKKK cascade |
| adrenergic, beta-2-, receptor, surface (ADRB2) | NM_000024.3 | 2.5 | activation of MAPK activity |
| mitogen-activated protein kinase 6 (MAPK6) | NM_002748.2 | 1.3 | MAP kinase activity |
| mitogen-activated protein kinase kinase kinase 6 (MAP3K6) | NM_004672.3 | 1.4 | protein serine/threonine kinase activity |
| mitogen-activated protein kinase kinase kinase 8 (MAP3K8) | NM_005204.2 | 11.1 | protein serine/threonine kinase activity |
| Inactivation | |||
| protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform (PPP2CA) | NM_002715.2 | 1.5 | inactivation of MAPK activity |
| dual specificity phosphatase 16 (DUSP16) | NM_030640.1 | 1.8 | inactivation of MAPK activity |
| dual specificity phosphatase 8 (DUSP8) | NM_004420.1 | 1.4 | inactivation of MAPK activity |
| RHOA | |||
| striated muscle activator of Rho-dependant signaling (STARS) | NM_139166.2 | 10.1 | positive regulation of Rho protein signal transduction |
| Rho family GTPase 3 (RND3) | NM_005168.3 | 5.4 | small GTPase mediated signal transduction |
| Rho guanine nucleotide exchange factor (GEF) 7 (ARHGEF7), transcript variant 2 | NM_145735.1 | 1.3 | RHOA positive regulation |
| Rho guanine nucleotide exchange factor (GEF) 12 (ARHGEF12) | NM_015313.1 | 1.2 | RHOA positive regulation |
| Rho GTPase activating protein 24 (ARHGAP24) | NM_031305.1 | 0.8 | RHOA negative regulation |
| Actin cytoskeleton and transcription factor activity | |||
| actinin, alpha 1 (ACTN1) | NM_001102.2 | 1.4 | actin cytoskeleton |
| diaphanous homolog 1 (Drosophila) (DIAPH1) | NM_005219.2 | 1.9 | actin cytoskeleton organization and biogenesis |
| coronin, actin binding protein, 1C (CORO1C) | NM_014325.2 | 1.5 | actin binding/cytoskeleton |
| filamin B, beta (actin binding protein 278) (FLNB) | NM_001457.1 | 1.5 | actin binding/cytoskeleton |
| actin, alpha 2, smooth muscle, aorta (ACTA2) | NM_001613.1 | 1.7 | actin filament |
| jun D proto-oncogene (JUND) | NM_005354.2 | 1.9 | activated transcription factor component |
| v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) | NM_005252.2 | 14.8 | activated transcription factor component |
| FBJ murine osteosarcoma viral oncogene homolog B (FOSB) | NM_006732.1 | 3.7 | activated transcription factor component |
| v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (MAFF), transcript variant 1 | NM_012323.2 | 2.6 | regulation of transcription |
| Other regulators of muscle growth and remodeling | |||
| activating transcription factor 3 (ATF3), transcript variant 3 | NM_001030287.1 | 28.4 | transcription factor activity |
| Xin actin-binding repeat containing 1 (XIRP1) | NM_194293.2 | 13.6 | actin cytoskeleton organization |
| v-myc myelocytomatosis viral oncogene homolog (avian) (MYC) | NM_002467.3 | 11.0 | transcription factor activity |
| heparin-binding EGF-like growth factor (HBEGF) | NM_001945.1 | 7.0 | growth factor activity |
| DnaJ (Hsp40) homolog, subfamily B, member 4 (DNAJB4) | NM_007034.3 | 6.1 | heat shock protein binding |
IPA identified several biological functions related to muscle growth and remodeling identified by the number of differentially expressed genes included in the function and the calculation of a P-value using a Fisher's Exact Test with a threshold for significance set at P≤0.05. They included: cancer (P<0.05, 233 molecules), gene expression (P<0.05, 146 molecules), cell assembly and organization (P<0.05, 69 molecules), cell morphology (P<0.05, 59 molecules) and skeletal and muscular system development and function (P<0.05, 50 molecules). In the same manner, IPA also identified three canonical pathways related to gene expression and growth/proliferation: ILK signaling (P = 0.005, 16/186 molecules), RAN signaling (P = 0.005, 4/23 molecules) and PI3K/AKT signaling (P = 0.006, 12/136 molecules).
RT-PCR analysis provides additional genes involved in actin dynamics and regulation of RhoA and NFAT signalling
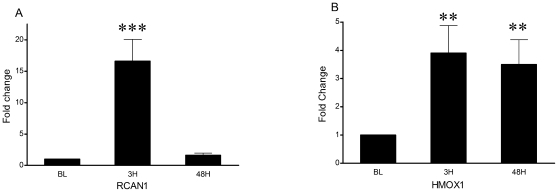
Targeted real time RT-PCR was conducted on several genes selected a priori for their involvement in recovery from skeletal muscle damage. Previous work using microarray analysis and RT-PCR identified the expression of novel genes following a similar eccentric protocol that are likely involved in the recovery and adaptation to damaging exercise [34]. Confirming the identification of two of its transcript variants in the microarray, RCAN1 mRNA content was highly elevated at 3H (16.6-fold, P<0.001) (Fig. 1A). Also induced was HMOX1 at both 3H (3.9-fold, P = 0.009) and 48H (3.5-fold, P = 0.002) (Fig. 1B).
Figure 1. Expression fold changes in mRNA expression of genes in muscle from baseline after exercise protocol.
Graph A – RCAN1 (N = 18). Graph B – HMOX1 (N = 18). 3H = 3 hours post exercise, 48H = 48 hours post exercise. Values are mean ± SEM. **Significant difference vs. baseline when collapsed across supplementation (P<0.01). ***Significant difference vs. baseline when collapsed across supplementation (P<0.001).
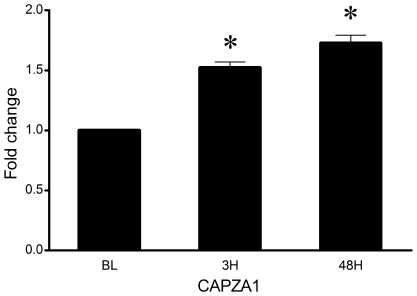
Expression of CAPZA1, a regulator of the growth of actin filaments, was increased at 48H (1.8-fold, P = 0.04) (Fig. 2).
Figure 2. Expression fold changes in mRNA expression of CAPZA1 in muscle from baseline after exercise protocol.
3H = 3 hours post exercise, 48H = 48 hours post exercise. N = 18. Values are mean ± SEM. *Significant difference vs. baseline when collapsed across supplementation (P<0.05).
Signaling proteins and protein content are affected by eccentric exercise
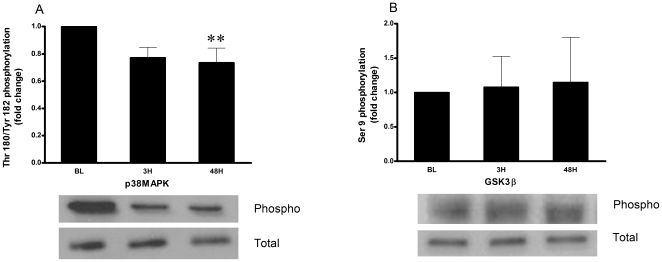
Phosphorylated p38MAPK and GSK-3β negatively regulate NFAT by promoting its export from the nucleus. Phosphorylation (Thr180/Tyr182) of p38MAPK was significantly lower at 3H (0.77-fold, P = 0.07) and 48H (0.73-fold, P = 0.005) as a result of exercise with no effect of E2 (Fig. 3A). Phosphorylation (Ser9) of GSK-3β was not affected by either exercise or E2 (Fig. 3B).
Figure 3. Fold change of phosphorylated/total ratio of signaling pathways from baseline after eccentric exercise.
BL = baseline, 3H = 3 hours post exercise, 48H = 48 hours post exercise. Graph A – p38MAPK (Thr 180/Tyr 182) (N = 18). Graph B – GSK-3β (Ser9) (N = 18). Values are mean ± SEM. **Significant difference vs. baseline when collapsed across supplementation (P<0.01).
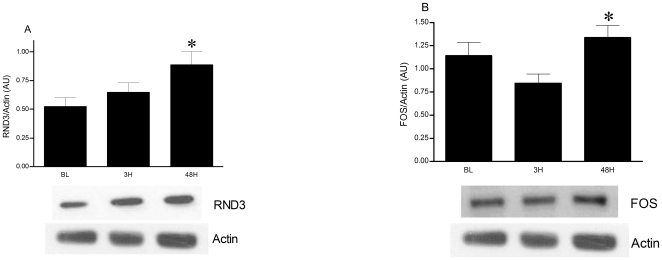
Two species highly expressed at 3H by the microarray were selected for western blot analysis. RND3 was significantly higher at both 3H (1.34-fold, P = 0.02) and 48H (1.39-fold, P<0.01) (Fig. 4A). FOS was significantly higher at 48 hours following exercise (1.16-fold, P<0.05) (Fig. 4B).
Figure 4. Western blot analysis of RND3 and FOS in skeletal muscle after eccentric exercise.
BL = baseline, 3H = 3 hours post exercise, 48H = 48 hours post exercise. Graph A – RND3 (N = 18). Graph B – FOS (N = 14). Values are mean ± SEM. *Significant difference vs. baseline when collapsed across supplementation (P<0.05).
Discussion
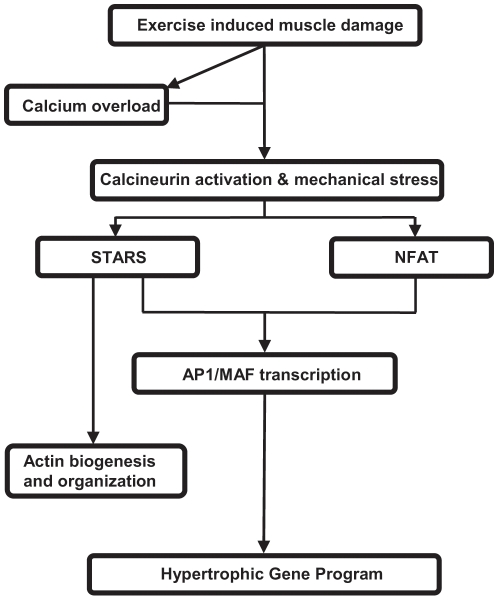
The sex hormone E2 displays anti-oxidant and membrane stabilizing properties that could protect skeletal muscle from the effects of exercise induced muscle damage and influence genetic expression patterns [50], [51]. Using microarray, real time RT-PCR and protein analyses, we have identified that 8 days of E2 supplementation did not affect the myofibre transcriptome in men. However, a single bout of eccentric exercise did induce differential mRNA transcription in the hypertrophic signaling pathways RhoA and nuclear factor of activated T-cells (NFAT) (Fig. 5), changes in the phosphorylation status of related signaling proteins and the protein quantities of two of the upregulated genes.
Figure 5. Schematic representation of the transcriptionally active pathways following exercise induced muscle damage.
Eccentric exercise promoted greater expression of targets within the STARS/RhoA/AP1 and NFAT/AP1 signaling pathways for hypertrophy and actin biogenesis and organization.
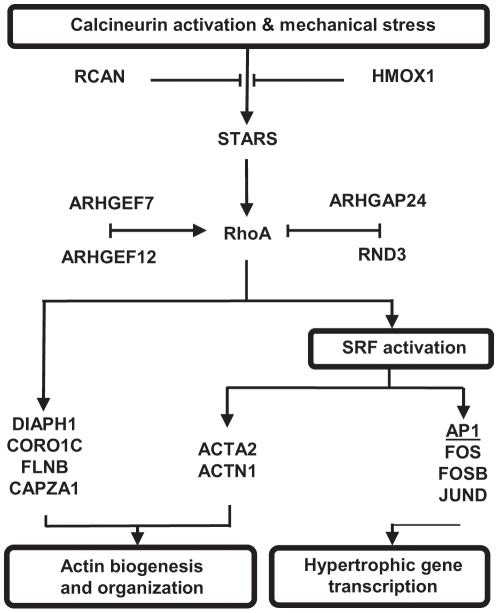
Of the genes affected early after exercise, one of the greatest inductions was observed in the novel actin binding protein striated muscle activator of Rho signaling (STARS). This protein is a muscle-specific transducer of cytoskeletal signaling in cardiac and skeletal muscle that responds to calcineurin activation and biomechanical stress [52], [53], [54], [55]. STARS stimulates growth through a mechanism requiring actin polymerization and Rho GTPase activation, increasing serum response factor (SRF)-mediated gene transcription (Fig. 6) [54], [55], [56], [57]. Originally identified in cardiac muscle, STARS mRNA content increases more than 3-fold following the hypertrophic signaling of pressure overload [52], [53]. More recently, Lamon et al. identified a 3.4-fold increase in STARS mRNA in human skeletal muscle following 8 weeks of resistance training [58]. Our measurement of more than a 10-fold increase suggests that this gene is very important for the early signaling for growth and remodeling following eccentric exercise.
Figure 6. Regulatory and downstream targets of STARS transcriptionally active following a single bout of eccentric exercise.
RCAN – regulator of calcineurin; HMOX1 – hemeoxygenase 1; ARHGEF7 and ARHGEF12 – Rho guanine nucleotide exchange factor 7 and 12; ARHGAP24 – Rho GTPase activating protein 24; RND3 – Rho family GTPase 3; DIAPH1 – diaphanous homologue 1; CORO1C – Coronin, actin binding protein 1; FLNB – Filamin B, beta; CAPZA1 – capping protein (actin filament) muscle Z-line alpha 1; ACTA2 – actin, alpha 2, smooth muscle, aorta; ACTN1 – actinin, alpha 1; AP1 – activator protein 1; FOS – FBJ murine osteosarcoma viral oncogene homologue; FOSB – FBJ murine osteosarcoma viral oncogene homologue B; JUND – Jun D proto-oncogene.
A downstream target of STARS associated with skeletal muscle hypertrophy and adaptation is the Rho GTPase, RhoA [58], [59]. Rho GTPases are a family of small signaling G proteins that interact with effector proteins to regulate actin cytoskeleton, cell cycle progression and gene transcription [3], [60], [61], [62]. These molecular signals switch to an active GTP bound state under the control of Rho GEFs (guanidine exchange factors), and return to their inactive GDP bound state by Rho GAPs (GTPase-activating proteins) [60], [63], . For the first time, our array analysis has identified increased expression of two GEFs (ARHGEF7 and ARHGEF12) and reduced expression of a GAP (ARHGAP24) that specifically regulate RhoA [60], [65]. These gene expression modifications, if translated into altered protein quantities, could increase the potential for RhoA activation.
Although increased activity of RhoA protein is necessary for myogenesis induction, it must be downregulated before myotube formation can proceed [66], [67]. This is achieved by RND3, another negative regulator of RhoA activity whose upregulation is an essential step of myoblast fusion [62], [68], [69]. Our damaging exercise protocol resulted in an early induction of this gene at 3H and elevated protein content by 48H. Cell culture experiments have identified that in the presence of growth factors, RND3 mRNA increases of ∼1.7-fold result in greater protein content within 30 h [62]. This in turn inhibits RhoA activity and promotes myotube formation and elongation [62].
Once activated, RhoA signaling is associated with myogenesis and actin remodeling in various cell types through regulation of genes that include DIAPH1 [70], [71], CORO1C [72], FLNB [73] and CAPZA1 [74]. Through SRF activation, RhoA and STARS also mediate the induction of actin proteins ACTA2 [75], ACTN1 [76] and members of the AP1 transcription factor complex: FOS [76], [77], FOSB [77] and JUND [78]. Each of these genes was induced by eccentric exercise at 3H, as identified by our gene array and targeted real time RT-PCR analysis and our data indicate that the increased transcription of FOS was effectively translated into protein, increasing levels significantly by 48H. Although the number of studies investigating these genes after exercise is few, some support can be found for the upregulation of select downstream targets. Following eccentric exercise, the largest induction occurs with the transcription factor FOS 2–8 hours post exercise (23 to 38-fold increases) [10], [79]. Resistance training results in a 2.7-fold increase in ACTN1 after 8 weeks [58]. Thirty minutes of high intensity running increased the expression of FOS (7.0-fold) FOSB (17.8-fold) and JUND (7.6-fold) [80]. Given that the mRNA levels for STARS, associated regulatory and transcription factors and downstream targeted genes were all significantly elevated 3 h after exercise, it appears that STARS signaling through a RhoA/SRF pathway is important for early skeletal muscle remodeling following damaging exercise.
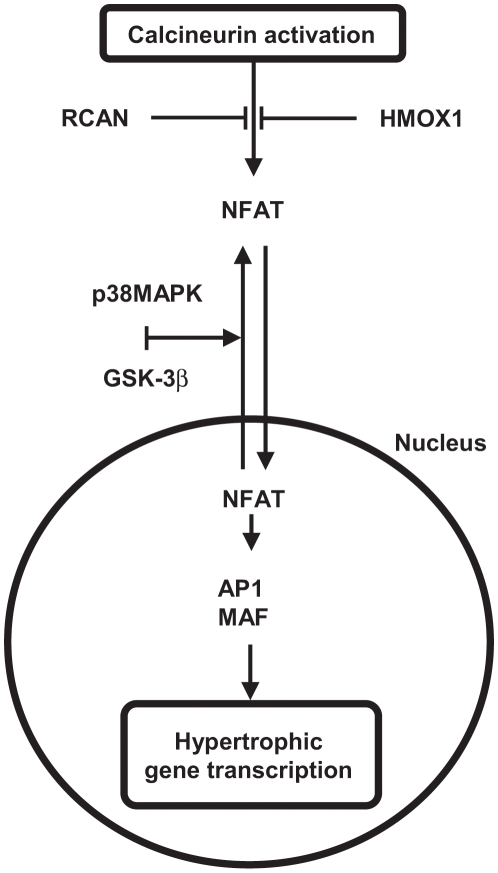
A second calcineurin influenced signaling pathway identified in our microarray analysis to be transcriptionally active was nuclear factor of activated T-cells (NFAT). NFAT proteins exist in the cytoplasm of cells in a phosphorylated and inactive state [81]. The influx of calcium following sustained contraction or damage increases the binding of calcineurin to NFAT, dephosphorylating conserved serine residues and promoting translocation of NFAT into the nucleus [81], [82], [83], [84]. Once inside the nucleus, NFAT cooperatively binds to DNA with transcription factors AP1 and MAF initiating the transcription of prohypertrophic genes (Fig. 7) [85]. Our microarray analysis also identified a significant induction of the genes NFATc1 and MAF at 3H. Along with the greater expression of the AP1 complex components FOS, FOSB and JUND, an increased abundance of NFATc1 and MAF could improve signaling by NFAT.
Figure 7. Regulatory and downstream targets of NFAT transcriptionally active following a single bout of eccentric exercise.
p38MAPK – p38 mitogen activated protein kinase; GSK-3β – glycogen synthase kinase 3 beta; MAF – v-maf musculoaponeurotic fibrosarcoma oncogene homologue (avian).
The NFAT pathway interacts with mitogen activated protein kinases (MAPK) and glycogen synthase kinase-3β (GSK-3β) for coordination of the hypertrophic response [84], [86]. p38MAPK and GSK-3β act as a negative regulators of cardiac hypertrophy, rephosphorylating NFAT and promoting its export from the nucleus [83], [86], [87]. Although GSK-3β mRNA content was higher in the microarray, its activity did not change and would not have affected NFAT nuclear residence. GoMiner analysis identified MAPK regulation as a transcriptionally active biological process through the induction of four related kinases (MAPKAPK2, MAPK6, MAP3K6 and MAP3K8), three related phosphatases (PP2CA, DUSP8 and DUSP16) and one surface receptor (ADRB2). Western blotting confirmed that p38MAPK phosphorylation status was lower after exercise, reaching significance by 48H. Lower activity of p38MAPK would assist in the transcriptional activity of NFAT, and may also occur to inhibit the induction of apoptosis and necrosis [2].
It should be noted that molecules that function in the recovery and repair of skeletal muscle through other mechanisms were also induced by our exercise protocol. Similar to other reports, ATF3 [10], MYC [10], [34], XIRP1 [88], HBEGF [10] and DNAJB4 [10], [34], [35] were highly up-regulated (6.1 to 28.4-fold) early after exercise. In addition, the altered mRNA content of members of the signaling pathways ILK, RAN and PI3K/AKT identified them as being transcriptionally active. Their respective functions in actin cytoskeleton remodeling [89], transport across the nuclear envelope for gene expression [90] and protein synthesis [91] relate to the top biological functions returned by gene ontology analysis. Interestingly, the biological function that contained the greatest number of molecules was cancer (233 molecules), likely due to similar alterations to the regulation of cellular growth experienced with both exercise and cancer.
Maintained muscle contractions and exercise-induced damage to the sarcoplasmic reticulum and sarcolemma may result in the accumulation of excess calcium, known as Ca2+ overload [5], [92]. As a signaling molecule, Ca2+ binds to calcineurin, activating both STARS [52] and NFAT [81], [82], [83], [84], [93]. Unrestrained calcineurin activity can be regulated in skeletal and cardiac muscle by the inhibitors regulator of calcineurin 1 (RCAN1, aka MCIP1, DSCR1) [81], [94], [95], [96], [97] and hemeoxygenase 1 (HMOX1) [98]. Increased mRNA content of RCAN1 was identified in microarray and targeted real time RT-PCR, confirming a previous gene expression profile that also identified an increase following exercise (3.8-fold) [34]. RT-PCR also identified significant increases in HMOX1 at both time points after exercise, similar to the 8-11-fold induction following 5 days of resistance training [99]. Together, the upregulation of these two calcineurin inhibitors identifies the importance of regulating the elevated calcineurin activity that occurred following unaccustomed eccentric exercise [97].
The primary mechanism through which estrogens influence the growth, differentiation and function of tissues occurs via the estrogen receptors ERα and ERβ [31], [32], [100]. As transcription factors, active homodimers and heterodimers of ERα and ERβ bind to estrogen response elements (EREs) in nuclear and mitochondrial DNA, increasing the transcription of target genes [101]. In a nongenomic manner, E2 also interacts with a number of proteins that include endothelial nitric oxide synthase (eNOS) and MAPK, altering the signals that modulate cellular differentiation, migration and survival [102], [103], [104]. Our observation that the myofibre transcriptome was unaffected by E2 is surprising. Although women have higher E2 concentrations than men, protein quantities of ERα and ERβ are similar and our supplementation protocol successfully increased circulating E2 to the level seen during the luteal phase of the menstrual cycle in healthy women [105], [106]. This suggests that the differential expression between men and women of genes involved in metabolism and growth regulation [36], [37], [38] may result from factors beyond E2 alone; factors that may include variations in body composition, X-chromosome genes, and/or other sex hormones (i.e., progesterone).
These results indicate that E2 supplementation does not affect the transcriptional pattern in skeletal muscle following eccentric exercise in men. However, the stress of a single bout of exercise induced a transcriptional response in two signaling pathways, STARS/RhoA/AP1 and NFAT/AP1, providing important insights for future research into the early hypertrophic response.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant in aid from the Natural Sciences and Engineering Council of Canada (NSERC) to M.T.(183817-07). S.M. was supported by financial assistance from the Glenn foundation for Medical Research. L.M. was supported by a graduate scholarship from NSERC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Favier FB, Benoit H, Freyssenet D. Cellular and molecular events controlling skeletal muscle mass in response to altered use. Pflugers Arch. 2008;456:587–600. doi: 10.1007/s00424-007-0423-z. [DOI] [PubMed] [Google Scholar]
- 2.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 3.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 4.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong RB. Initial events in exercise-induced muscular injury. Med Sci Sports Exerc. 1990;22:429–435. [PubMed] [Google Scholar]
- 6.Clarkson PM, Hubal MJ. Exercise-induced muscle damage in humans. Am J Phys Med Rehabil. 2002;81:S52–69. doi: 10.1097/00002060-200211001-00007. [DOI] [PubMed] [Google Scholar]
- 7.Newham DJ, Jones DA, Clarkson PM. Repeated high-force eccentric exercise: effects on muscle pain and damage. J Appl Physiol. 1987;63:1381–1386. doi: 10.1152/jappl.1987.63.4.1381. [DOI] [PubMed] [Google Scholar]
- 8.Jones DA, Newham DJ, Torgan C. Mechanical influences on long-lasting human muscle fatigue and delayed-onset pain. J Physiol. 1989;412:415–427. doi: 10.1113/jphysiol.1989.sp017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friden J, Lieber RL. Eccentric exercise-induced injuries to contractile and cytoskeletal muscle fibre components. Acta Physiol Scand. 2001;171:321–326. doi: 10.1046/j.1365-201x.2001.00834.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol. 2003;95:2485–2494. doi: 10.1152/japplphysiol.01161.2002. [DOI] [PubMed] [Google Scholar]
- 11.Arnett MG, Hyslop R, Dennehy CA, Schneider CM. Age-related variations of serum CK and CK MB response in females. Can J Appl Physiol. 2000;25:419–429. doi: 10.1139/h00-027. [DOI] [PubMed] [Google Scholar]
- 12.Beaton LJ, Tarnopolsky MA, Phillips SM. Contraction-induced muscle damage in humans following calcium channel blocker administration. J Physiol. 2002;544:849–859. doi: 10.1113/jphysiol.2002.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friden J, Kjorell U, Thornell LE. Delayed muscle soreness and cytoskeletal alterations: an immunocytological study in man. Int J Sports Med. 1984;5:15–18. doi: 10.1055/s-2008-1025873. [DOI] [PubMed] [Google Scholar]
- 14.Newham DJ, McPhail G, Mills KR, Edwards RH. Ultrastructural changes after concentric and eccentric contractions of human muscle. J Neurol Sci. 1983;61:109–122. doi: 10.1016/0022-510x(83)90058-8. [DOI] [PubMed] [Google Scholar]
- 15.Amelink GJ, Kamp HH, Bar PR. Creatine kinase isoenzyme profiles after exercise in the rat: sex-linked differences in leakage of CK-MM. Pflugers Arch. 1988;412:417–421. doi: 10.1007/BF01907561. [DOI] [PubMed] [Google Scholar]
- 16.Stupka N, Tarnopolsky MA, Yardley NJ, Phillips SM. Cellular adaptation to repeated eccentric exercise-induced muscle damage. J Appl Physiol. 2001;91:1669–1678. doi: 10.1152/jappl.2001.91.4.1669. [DOI] [PubMed] [Google Scholar]
- 17.Amelink GJ, Koot RW, Erich WB, Van Gijn J, Bar PR. Sex-linked variation in creatine kinase release, and its dependence on oestradiol, can be demonstrated in an in-vitro rat skeletal muscle preparation. Acta Physiol Scand. 1990;138:115–124. doi: 10.1111/j.1748-1716.1990.tb08823.x. [DOI] [PubMed] [Google Scholar]
- 18.Amelink GJ, Bar PR. Exercise-induced muscle protein leakage in the rat. Effects of hormonal manipulation. J Neurol Sci. 1986;76:61–68. doi: 10.1016/0022-510x(86)90142-5. [DOI] [PubMed] [Google Scholar]
- 19.Stupka N, Tiidus PM. Effects of ovariectomy and estrogen on ischemia-reperfusion injury in hindlimbs of female rats. J Appl Physiol. 2001;91:1828–1835. doi: 10.1152/jappl.2001.91.4.1828. [DOI] [PubMed] [Google Scholar]
- 20.Komulainen J, Koskinen SO, Kalliokoski R, Takala TE, Vihko V. Gender differences in skeletal muscle fibre damage after eccentrically biased downhill running in rats. Acta Physiol Scand. 1999;165:57–63. doi: 10.1046/j.1365-201x.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 21.Enns DL, Iqbal S, Tiidus PM. Oestrogen receptors mediate oestrogen-induced increases in post-exercise rat skeletal muscle satellite cells. Acta Physiol (Oxf) 2008;194:81–93. doi: 10.1111/j.1748-1716.2008.01861.x. [DOI] [PubMed] [Google Scholar]
- 22.Tiidus PM, Holden D, Bombardier E, Zajchowski S, Enns D, et al. Estrogen effect on post-exercise skeletal muscle neutrophil infiltration and calpain activity. Can J Physiol Pharmacol. 2001;79:400–406. [PubMed] [Google Scholar]
- 23.Stupka N, Lowther S, Chorneyko K, Bourgeois JM, Hogben C, et al. Gender differences in muscle inflammation after eccentric exercise. J Appl Physiol. 2000;89:2325–2332. doi: 10.1152/jappl.2000.89.6.2325. [DOI] [PubMed] [Google Scholar]
- 24.St Pierre Schneider B, Correia LA, Cannon JG. Sex differences in leukocyte invasion in injured murine skeletal muscle. Res Nurs Health. 1999;22:243–250. doi: 10.1002/(sici)1098-240x(199906)22:3<243::aid-nur6>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 25.Tiidus PM, Deller M, Liu XL. Oestrogen influence on myogenic satellite cells following downhill running in male rats: a preliminary study. Acta Physiol Scand. 2005;184:67–72. doi: 10.1111/j.1365-201X.2005.01427.x. [DOI] [PubMed] [Google Scholar]
- 26.Enns DL, Tiidus PM. Estrogen influences satellite cell activation and proliferation following downhill running in rats. J Appl Physiol. 2008;104:347–353. doi: 10.1152/japplphysiol.00128.2007. [DOI] [PubMed] [Google Scholar]
- 27.Persky AM, Green PS, Stubley L, Howell CO, Zaulyanov L, et al. Protective effect of estrogens against oxidative damage to heart and skeletal muscle in vivo and in vitro. Proc Soc Exp Biol Med. 2000;223:59–66. doi: 10.1046/j.1525-1373.2000.22308.x. [DOI] [PubMed] [Google Scholar]
- 28.Sribnick EA, Ray SK, Banik NL. Estrogen as a multi-active neuroprotective agent in traumatic injuries. Neurochem Res. 2004;29:2007–2014. doi: 10.1007/s11064-004-6874-0. [DOI] [PubMed] [Google Scholar]
- 29.Tiidus PM. Can estrogens diminish exercise induced muscle damage? Can J Appl Physiol. 1995;20:26–38. doi: 10.1139/h95-002. [DOI] [PubMed] [Google Scholar]
- 30.Kahlert S, Grohe C, Karas RH, Lobbert K, Neyses L, et al. Effects of estrogen on skeletal myoblast growth. Biochem Biophys Res Commun. 1997;232:373–378. doi: 10.1006/bbrc.1997.6223. [DOI] [PubMed] [Google Scholar]
- 31.Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. N Engl J Med. 2002;346:340–352. doi: 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- 32.Katzenellenbogen BS, Choi I, Delage-Mourroux R, Ediger TR, Martini PG, et al. Molecular mechanisms of estrogen action: selective ligands and receptor pharmacology. J Steroid Biochem Mol Biol. 2000;74:279–285. doi: 10.1016/s0960-0760(00)00104-7. [DOI] [PubMed] [Google Scholar]
- 33.Clarkson PM, Hubal MJ. Are women less susceptible to exercise-induced muscle damage? Curr Opin Clin Nutr Metab Care. 2001;4:527–531. doi: 10.1097/00075197-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 34.Mahoney DJ, Safdar A, Parise G, Melov S, Fu M, et al. Gene expression profiling in human skeletal muscle during recovery from eccentric exercise. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1901–1910. doi: 10.1152/ajpregu.00847.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, et al. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4:R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maher AC, Fu MH, Isfort RJ, Varbanov AR, Qu XA, et al. Sex differences in global mRNA content of human skeletal muscle. PLoS One. 2009;4:e6335. doi: 10.1371/journal.pone.0006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welle S, Tawil R, Thornton CA. Sex-related differences in gene expression in human skeletal muscle. PLoS One. 2008;3:e1385. doi: 10.1371/journal.pone.0001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth SM, Ferrell RE, Peters DG, Metter EJ, Hurley BF, et al. Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genomics. 2002;10:181–190. doi: 10.1152/physiolgenomics.00028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Housh DJ, Housh TJ, Weir JP, Weir LL, Johnson GO, et al. Anthropometric estimation of thigh muscle cross-sectional area. Med Sci Sports Exerc. 1995;27:784–791. [PubMed] [Google Scholar]
- 40.Devries MC, Hamadeh MJ, Graham TE, Tarnopolsky MA. 17beta-estradiol supplementation decreases glucose rate of appearance and disappearance with no effect on glycogen utilization during moderate intensity exercise in men. J Clin Endocrinol Metab. 2005;90:6218–6225. doi: 10.1210/jc.2005-0926. [DOI] [PubMed] [Google Scholar]
- 41.Beaton LJ, Allan DA, Tarnopolsky MA, Tiidus PM, Phillips SM. Contraction-induced muscle damage is unaffected by vitamin E supplementation. Med Sci Sports Exerc. 2002;34:798–805. doi: 10.1097/00005768-200205000-00012. [DOI] [PubMed] [Google Scholar]
- 42.Vissing K, Andersen JL, Schjerling P. Are exercise-induced genes induced by exercise? Faseb J. 2005;19:94–96. doi: 10.1096/fj.04-2084fje. [DOI] [PubMed] [Google Scholar]
- 43.Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, et al. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18:226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- 44.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu XM, Peyton KJ, Ensenat D, Wang H, Schafer AI, et al. Endoplasmic reticulum stress stimulates heme oxygenase-1 gene expression in vascular smooth muscle. Role in cell survival. J Biol Chem. 2005;280:872–877. doi: 10.1074/jbc.M410413200. [DOI] [PubMed] [Google Scholar]
- 46.Melov S, Tarnopolsky MA, Beckman K, Felkey K, Hubbard A. Resistance exercise reverses aging in human skeletal muscle. PLoS ONE. 2007;2:e465. doi: 10.1371/journal.pone.0000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 48.Tarnopolsky MA, Parshad A, Walzel B, Schlattner U, Wallimann T. Creatine transporter and mitochondrial creatine kinase protein content in myopathies. Muscle Nerve. 2001;24:682–688. doi: 10.1002/mus.1055. [DOI] [PubMed] [Google Scholar]
- 49.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 50.Tiidus PM, Enns DL. Point:Counterpoint: Estrogen and sex do/do not influence post-exercise indexes of muscle damage, inflammation, and repair. J Appl Physiol. 2009;106:1010–1012; discussion 1014–1015, 1021. doi: 10.1152/japplphysiol.90848.2008. [DOI] [PubMed] [Google Scholar]
- 51.Tiidus PM. Can oestrogen influence skeletal muscle damage, inflammation, and repair? Br J Sports Med. 2005;39:251–253. doi: 10.1136/bjsm.2005.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuwahara K, Teg Pipes GC, McAnally J, Richardson JA, Hill JA, et al. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117:1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahadeva H, Brooks G, Lodwick D, Chong NW, Samani NJ. ms1, a novel stress-responsive, muscle-specific gene that is up-regulated in the early stages of pressure overload-induced left ventricular hypertrophy. FEBS Lett. 2002;521:100–104. doi: 10.1016/s0014-5793(02)02833-8. [DOI] [PubMed] [Google Scholar]
- 54.Arai A, Spencer JA, Olson EN. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J Biol Chem. 2002;277:24453–24459. doi: 10.1074/jbc.M202216200. [DOI] [PubMed] [Google Scholar]
- 55.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrientos T, Frank D, Kuwahara K, Bezprozvannaya S, Pipes GC, et al. Two novel members of the ABLIM protein family, ABLIM-2 and -3, associate with STARS and directly bind F-actin. J Biol Chem. 2007;282:8393–8403. doi: 10.1074/jbc.M607549200. [DOI] [PubMed] [Google Scholar]
- 57.Wei L, Zhou W, Croissant JD, Johansen FE, Prywes R, et al. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J Biol Chem. 1998;273:30287–30294. doi: 10.1074/jbc.273.46.30287. [DOI] [PubMed] [Google Scholar]
- 58.Lamon S, Wallace MA, Leger B, Russell AP. Regulation of STARS and its downstream targets suggest a novel pathway involved in human skeletal muscle hypertrophy and atrophy. J Physiol. 2009;587:1795–1803. doi: 10.1113/jphysiol.2009.168674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takano H, Komuro I, Oka T, Shiojima I, Hiroi Y, et al. The Rho family G proteins play a critical role in muscle differentiation. Mol Cell Biol. 1998;18:1580–1589. doi: 10.1128/mcb.18.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 61.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 62.Fortier M, Comunale F, Kucharczak J, Blangy A, Charrasse S, et al. RhoE controls myoblast alignment prior fusion through RhoA and ROCK. Cell Death Differ. 2008;15:1221–1231. doi: 10.1038/cdd.2008.34. [DOI] [PubMed] [Google Scholar]
- 63.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 64.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 65.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 66.Charrasse S, Comunale F, Grumbach Y, Poulat F, Blangy A, et al. RhoA GTPase regulates M-cadherin activity and myoblast fusion. Mol Biol Cell. 2006;17:749–759. doi: 10.1091/mbc.E05-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castellani L, Salvati E, Alema S, Falcone G. Fine regulation of RhoA and Rock is required for skeletal muscle differentiation. J Biol Chem. 2006;281:15249–15257. doi: 10.1074/jbc.M601390200. [DOI] [PubMed] [Google Scholar]
- 68.Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, et al. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol. 2003;13:1106–1115. doi: 10.1016/s0960-9822(03)00418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol. 1998;18:4761–4771. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Y, Tan EJ, Wee S, Manser E, Lim L, et al. Functional interactions between phosphatase POPX2 and mDia modulate RhoA pathways. J Cell Sci. 2008;121:514–521. doi: 10.1242/jcs.013557. [DOI] [PubMed] [Google Scholar]
- 71.Gao G, Chen L, Dong B, Gu H, Dong H, et al. RhoA effector mDia1 is required for PI 3-kinase-dependent actin remodeling and spreading by thrombin in platelets. Biochem Biophys Res Commun. 2009;385:439–444. doi: 10.1016/j.bbrc.2009.05.090. [DOI] [PubMed] [Google Scholar]
- 72.Xavier CP, Rastetter RH, Stumpf M, Rosentreter A, Muller R, et al. Structural and Functional Diversity of Novel Coronin 1C (CRN2) Isoforms in Muscle. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 73.Bello NF, Lamsoul I, Heuze ML, Metais A, Moreaux G, et al. The E3 ubiquitin ligase specificity subunit ASB2beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ. 2009;16:921–932. doi: 10.1038/cdd.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papa I, Astier C, Kwiatek O, Raynaud F, Bonnal C, et al. Alpha actinin-CapZ, an anchoring complex for thin filaments in Z-line. J Muscle Res Cell Motil. 1999;20:187–197. doi: 10.1023/a:1005489319058. [DOI] [PubMed] [Google Scholar]
- 75.Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, et al. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci U S A. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson TJ, Balza R, Jr, Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J Mol Cell Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 77.Santalucia T, Christmann M, Yacoub MH, Brand NJ. Hypertrophic agonists induce the binding of c-Fos to an AP-1 site in cardiac myocytes: implications for the expression of GLUT1. Cardiovasc Res. 2003;59:639–648. doi: 10.1016/s0008-6363(03)00472-3. [DOI] [PubMed] [Google Scholar]
- 78.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 79.Trenerry MK, Carey KA, Ward AC, Cameron-Smith D. STAT3 signaling is activated in human skeletal muscle following acute resistance exercise. J Appl Physiol. 2007;102:1483–1489. doi: 10.1152/japplphysiol.01147.2006. [DOI] [PubMed] [Google Scholar]
- 80.Puntschart A, Wey E, Jostarndt K, Vogt M, Wittwer M, et al. Expression of fos and jun genes in human skeletal muscle after exercise. Am J Physiol. 1998;274:C129–137. doi: 10.1152/ajpcell.1998.274.1.C129. [DOI] [PubMed] [Google Scholar]
- 81.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 82.Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun. 2004;322:1178–1191. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- 83.Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 84.Fiedler B, Wollert KC. Interference of antihypertrophic molecules and signaling pathways with the Ca2+-calcineurin-NFAT cascade in cardiac myocytes. Cardiovasc Res. 2004;63:450–457. doi: 10.1016/j.cardiores.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 85.Crabtree GR. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 86.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 87.Braz JC, Bueno OF, Liang Q, Wilkins BJ, Dai YS, et al. Targeted inhibition of p38 MAPK promotes hypertrophic cardiomyopathy through upregulation of calcineurin-NFAT signaling. J Clin Invest. 2003;111:1475–1486. doi: 10.1172/JCI17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hawke TJ, Atkinson DJ, Kanatous SB, Van der Ven PF, Goetsch SC, et al. Xin, an actin binding protein, is expressed within muscle satellite cells and newly regenerated skeletal muscle fibers. Am J Physiol Cell Physiol. 2007;293:C1636–1644. doi: 10.1152/ajpcell.00124.2007. [DOI] [PubMed] [Google Scholar]
- 89.Postel R, Vakeel P, Topczewski J, Knoll R, Bakkers J. Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev Biol. 2008;318:92–101. doi: 10.1016/j.ydbio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 90.Kardassis D, Murphy C, Fotsis T, Moustakas A, Stournaras C. Control of transforming growth factor beta signal transduction by small GTPases. Febs J. 2009;276:2947–2965. doi: 10.1111/j.1742-4658.2009.07031.x. [DOI] [PubMed] [Google Scholar]
- 91.Sasai N, Agata N, Inoue-Miyazu M, Kawakami K, Kobayashi K, et al. Involvement of PI3K/Akt/TOR pathway in stretch-induced hypertrophy of myotubes. Muscle Nerve. 2009 doi: 10.1002/mus.21473. [DOI] [PubMed] [Google Scholar]
- 92.Wright DC. Mechanisms of calcium-induced mitochondrial biogenesis and GLUT4 synthesis. Appl Physiol Nutr Metab. 2007;32:840–845. doi: 10.1139/H07-062. [DOI] [PubMed] [Google Scholar]
- 93.Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 94.Rothermel BA, Vega RB, Williams RS. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med. 2003;13:15–21. doi: 10.1016/s1050-1738(02)00188-3. [DOI] [PubMed] [Google Scholar]
- 95.Oh M, Rybkin II, Copeland V, Czubryt MP, Shelton JM, et al. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol Cell Biol. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leinwand LA. Calcineurin inhibition and cardiac hypertrophy: a matter of balance. Proc Natl Acad Sci U S A. 2001;98:2947–2949. doi: 10.1073/pnas.051033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang J, Rothermel B, Vega RB, Frey N, McKinsey TA, et al. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ Res. 2000;87:E61–68. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- 98.Tongers J, Fiedler B, Konig D, Kempf T, Klein G, et al. Heme oxygenase-1 inhibition of MAP kinases, calcineurin/NFAT signaling, and hypertrophy in cardiac myocytes. Cardiovasc Res. 2004;63:545–552. doi: 10.1016/j.cardiores.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 99.Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am J Physiol Endocrinol Metab. 2000;279:E806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- 100.Mendez P, Azcoitia I, Garcia-Segura LM. Interdependence of oestrogen and insulin-like growth factor-I in the brain: potential for analysing neuroprotective mechanisms. J Endocrinol. 2005;185:11–17. doi: 10.1677/joe.1.06058. [DOI] [PubMed] [Google Scholar]
- 101.Klinge CM. Estrogenic control of mitochondrial function and biogenesis. J Cell Biochem. 2008;105:1342–1351. doi: 10.1002/jcb.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009;20:477–482. doi: 10.1016/j.tem.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim HP, Lee JY, Jeong JK, Bae SW, Lee HK, et al. Nongenomic stimulation of nitric oxide release by estrogen is mediated by estrogen receptor alpha localized in caveolae. Biochem Biophys Res Commun. 1999;263:257–262. doi: 10.1006/bbrc.1999.1348. [DOI] [PubMed] [Google Scholar]
- 104.Watters JJ, Campbell JS, Cunningham MJ, Krebs EG, Dorsa DM. Rapid membrane effects of steroids in neuroblastoma cells: effects of estrogen on mitogen activated protein kinase signalling cascade and c-fos immediate early gene transcription. Endocrinology. 1997;138:4030–4033. doi: 10.1210/endo.138.9.5489. [DOI] [PubMed] [Google Scholar]
- 105.Wiik A, Glenmark B, Ekman M, Esbjornsson-Liljedahl M, Johansson O, et al. Oestrogen receptor beta is expressed in adult human skeletal muscle both at the mRNA and protein level. Acta Physiol Scand. 2003;179:381–387. doi: 10.1046/j.0001-6772.2003.01186.x. [DOI] [PubMed] [Google Scholar]
- 106.Wiik A, Ekman M, Johansson O, Jansson E, Esbjornsson M. Expression of both oestrogen receptor alpha and beta in human skeletal muscle tissue. Histochem Cell Biol. 2009;131:181–189. doi: 10.1007/s00418-008-0512-x. [DOI] [PubMed] [Google Scholar]