Summary
We have profiled promoter DNA methylation alterations in 272 glioblastoma tumors in the context of The Cancer Genome Atlas (TCGA). We found that a distinct subset of samples displays concerted hypermethylation at a large number of loci, indicating the existence of a glioma-CpG Island Methylator Phenotype (G-CIMP). We validated G-CIMP in a set of non-TCGA glioblastomas and low-grade gliomas. G-CIMP tumors belong to the Proneural subgroup, are more prevalent among low-grade gliomas, display distinct copy-number alterations and are tightly associated with IDH1 somatic mutations. Patients with G-CIMP tumors are younger at the time of diagnosis and experience significantly improved outcome. These findings identify G-CIMP as a distinct subset of human gliomas on molecular and clinical grounds.
Keywords: DNA methylation, glioma, CIMP, IDH1, TCGA
Introduction
Human gliomas present as heterogeneous disease, primarily defined by the histologic appearance of the tumor cells. Astrocytoma and oligodendroglioma constitute the infiltrating gliomas (Adamson et al., 2009). The cell or cells of origin have not definitively been identified, however, the identification of tumorigenic, stem-cell like precursor cells in advanced stage gliomas suggests that human gliomas may have a neural stem cell origin (Canoll and Goldman, 2008; Dirks, 2006; Galli et al., 2004).
Gliomas are subdivided by the World Health Organization (WHO) by histological grade, which is an indication of differentiation status, malignant potential, response to treatment and survival. Glioblastoma (GBM), also described as Grade IV glioma, accounts for more than 50% of all gliomas (Adamson et al., 2009). Patients with GBM have an overall median survival time of only 15 months (Brandes et al., 2001; Martinez et al., 2010; Parsons et al., 2008). Most GBMs are diagnosed as de novo, or primary tumors and are more common in males. A subset of ~5% of GBM tumors, termed secondary GBM, progress from lower grade tumors (grade II/III), are seen in younger patients, are more evenly distributed among the sexes and exhibit longer survival times (reviewed in (Adamson et al., 2009; Furnari et al., 2007).
There is currently great interest in characterizing and compiling the genome and transcriptome changes in human GBM tumors to identify aberrantly functioning molecular pathways and tumor subtypes. The Cancer Genome Atlas (TCGA) pilot project identified genetic changes of primary DNA sequence and copy number, DNA methylation, gene expression and patient clinical information for a set of GBM tumors (The Cancer Genome Atlas Research Network, 2008). TCGA also reaffirmed genetic alterations in TP53, PTEN, EGFR, RB1 NF1, ERBB2, PIK3R1, and PIK3CA mutations and detected an increased frequency of NF1 mutations in GBM patients (The Cancer Genome Atlas Research Network, 2008). Recent DNA sequencing analyses of primary GBM tumors using a more comprehensive approach (Parsons et al., 2008) also identified novel somatic mutations in isocitrate dehydrogenase 1 (IDH1) that occur in 12% of all GBM patients. IDH1 mutations have only been detected at the arginine residue in codon 132, with the most common change being the R132H mutation (Parsons et al., 2008; Yan et al., 2009), which results in a novel gain of enzyme function in directly catalyzing α-ketoglutarate to R(−)-2-hydroxyglutarate (Dang et al., 2009). IDH1 mutations are enriched in secondary GBM cases, younger individuals and coincident with increased patient survival (Balss et al., 2008; Hartmann et al., 2009; Yan et al., 2009). Higher IDH1 mutation rates are seen in grade II and III astrocytomas and oligodendrogliomas (Balss et al., 2008; Bleeker et al., 2009; Hartmann et al., 2009; Yan et al., 2009), suggesting that IDH1 mutations generally occur in the progressive form of glioma, rather than in de novo GBM. Mutations in the related IDH2 gene are of lower frequency and generally non-overlapping with tumors containing IDH1 mutations (Hartmann et al., 2009; Yan et al., 2009).
Cancer-specific DNA methylation changes are hallmarks of human cancers, in which global DNA hypomethylation is often seen concomitantly with hypermethylation of CpG islands (reviewed in (Jones and Baylin, 2007)). Promoter CpG island hypermethylation generally results in transcriptional silencing of the associated gene (Jones and Baylin, 2007). CpG island hypermethylation events have also been shown to serve as biomarkers in human cancers, for early detection in blood and other bodily fluids, for prognosis or prediction of response to therapy and to monitor cancer recurrence (Laird, 2003).
A CpG island methylator phenotype (CIMP) was first characterized in human colorectal cancer by Toyota and colleagues (Toyota et al., 1999) as cancer-specific CpG island hypermethylation of a subset of genes in a subset of tumors. We confirmed and further characterized colorectal CIMP using MethyLight technology (Weisenberger et al., 2006). Colorectal CIMP is characterized by tumors in the proximal colon, a tight association with BRAF mutations, and microsatellite instability caused by MLH1 promoter hypermethylation and transcriptional silencing (Weisenberger et al., 2006).
DNA methylation alterations have been widely reported in human gliomas, and there have been several reports of promoter-associated CpG island hypermethylation in human GBM and other glioma subtypes (Kim et al., 2006; Martinez et al., 2009; Martinez et al., 2007; Nagarajan and Costello, 2009; Stone et al., 2004; Tepel et al., 2008; Uhlmann et al., 2003). Several studies have noted differences between primary and secondary GBMs with respect to epigenetic changes. Overall secondary GBMs have a higher frequency of promoter methylation than primary GBM (Ohgaki and Kleihues, 2007). In particular, promoter methylation of RB1 was found to be approximately three times more common in secondary GBM (Nakamura et al., 2001).
Hypermethylation of the MGMT promoter-associated CpG island has been shown in a large percentage of GBM patients (Esteller et al., 2000; Esteller et al., 1999; Hegi et al., 2005; Hegi et al., 2008; Herman and Baylin, 2003). MGMT encodes for an O6-methylguanine methyltransferase which removes alkyl groups from the O-6 position of guanine. GBM patients with MGMT hypermethylation showed sensitivity to alkyating agents such as temozolomide, with an accompanying improved outcome (Esteller et al., 2000; Esteller et al., 1999; Hegi et al., 2005; Hegi et al., 2008; Herman and Baylin, 2003). However, initial promoter methylation of MGMT, in conjunction with temozolomide treatment, may result in selective pressure to lose mismatch repair function, resulting in aggressive recurrent tumors with a hypermutator phenotype (Cahill et al., 2007; Hegi et al., 2005; Silber et al., 1999; The Cancer Genome Atlas Research Network, 2008).
Here we report the DNA methylation analysis of 272 GBM tumors collected for TCGA, extend this to lower grade tumors and characterize a distinct subgroup of human gliomas exhibiting CIMP.
Results
Identification of a distinct DNA methylation subgroup within GBM patients
We determined DNA methylation profiles in a discovery set of 272 TCGA GBM samples. At the start of this study, we relied on the Illumina GoldenGate platform, using both the standard Cancer Panel I, and a custom-designed array (The Cancer Genome Atlas Research Network, 2008) (Figure S1A–C, Experimental Procedures), but migrated to the more comprehensive Infinium platform (Figure 1, Figure S1C), as it became available. DNA methylation measurements were highly correlated for CpG dinucleotides shared between the two platforms (Pearson’s r = 0.94, Figure S1D). Both platforms interrogate a sampling of about two CpG dinucleotides per gene. Although this implies that non-representative CpGs may be assessed for some promoters, it is likely that representative results will be obtained for most gene promoters, given the very high degree of locally correlated DNA methylation behavior (Eckhardt et al., 2006).
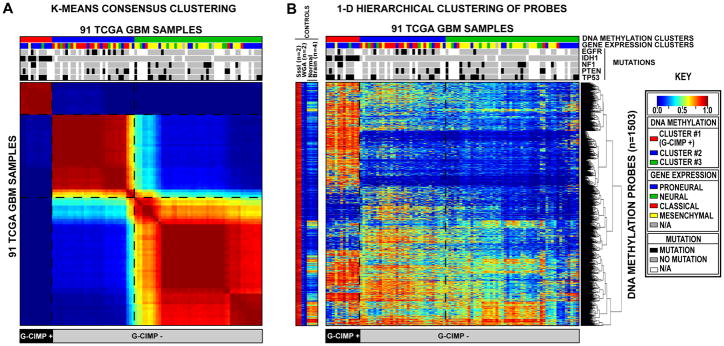
Figure 1. Clustering of TCGA GBM tumors and control samples identifies a CpG Island Methylator Phenotype (G-CIMP).
Unsupervised consensus clustering was performed using the 1,503 Infnium DNA methylation probes whose DNA methylation beta values varied the most across the 91 TCGA GBM samples. DNA Methylation clusters are distinguished with a color code at the top of the panel: red, consensus cluster 1 (n=12 tumors); blue, consensus cluster 2 (n=31 tumors); green, consensus cluster 3 (n=48 samples). Each sample within each DNA Methylation cluster are colored labeled as described in the key for its gene expression cluster membership (Proneural, Neural, Classical and Mesenchymal). The somatic mutation status of five genes (EGFR, IDH1, NF1, PTEN, and TP53) are indicated by the black squares, the gray squares indicate the absence of mutations in the sample and the white squares indicate that the gene was not screened in the specific sample. G-CIMP-positive samples are labeled at the bottom of the matrix. A) Consensus matrix produced by k-means clustering (K = 3). The samples are listed in the same order on the x and y axes. Consensus index values range from 0 to 1, 0 being highly dissimilar and 1 being highly similar. B) One-dimensional hierarchical clustering of the same 1,503 most variant probes, with retention of the same sample order as in Figure 1A. Each row represents a probe; each column represents a sample. The level of DNA methylation (beta value) for each probe, in each sample, is represented by using a color scale as shown in the legend; white indicates missing data. M.SssI-treated DNA (n=2), WGA-DNA (n=2) and normal brain (n=4) samples are included in the heatmap but did not contribute to the unsupervised clustering. The probes in the eight control samples are listed in the same order as the y axis of the GBM sample heatmap. See also Figure S1 and Table S1.
We selected the most variant probes on each platform and performed consensus clustering to identify GBM subgroups (Monti et al., 2003). We identified three DNA methylation clusters using either the GoldenGate or Infinium data, with 97% concordance (61/63) in cluster membership calls for samples run on both platforms (Table S1). Cluster 1 formed a particularly tight cluster on both platforms with a highly characteristic DNA methylation profile (Figure 1, Figure S1), reminiscent of the CpG Island Methylator Phenotype described in colorectal cancer (Toyota et al., 1999; Weisenberger et al., 2006). CIMP in colorectal cancer is characterized by correlated cancer-specific CpG island hypermethylation of a subset of genes in a subset of tumors, and not just a stochastic increase in the frequency of generic CpG island methylation across the genome (Toyota et al., 1999; Weisenberger et al., 2006). Cluster 1 GBM samples show similar concerted methylation changes at a subset of loci. We therefore designated cluster 1 tumors as having a glioma CpG Island Methylator Phenotype (G-CIMP). Combining Infinium and GoldenGate data, 24 of 272 TCGA GBM samples (8.8%) were identified as G-CIMP subtype (Table S1).
Characterization of G-CIMP tumors within gene expression clusters
Four gene expression subtypes (Proneural, Neural, Classical and Mesenchymal) have been previously identified and characterized using TCGA GBM samples (Verhaak et al., 2010). We compared the DNA methylation consensus cluster assignments for each sample to their gene expression cluster assignments (Figure 1, Figure 2A, Figure S1A–B, Table S1). The G-CIMP sample cluster is highly enriched for proneural GBM tumors, while the DNA methylation clusters 2 and 3 are moderately enriched for classical and mesenchymal expression groups, respectively. Of the 24 G-CIMP tumors, 21 (87.5%) were classified within the proneural expression group. These G-CIMP tumors represent 30% (21/71) of all proneural GBM tumors, suggesting that G-CIMP tumors represent a distinct subset of proneural GBM tumors (Figure 2A and Figure S2A, Table S1). The few non-proneural G-CIMP tumors belong to neural (2/24 tumors, 8.3%), and mesenchymal (1/24 tumors, 4.2%) gene expression groups.
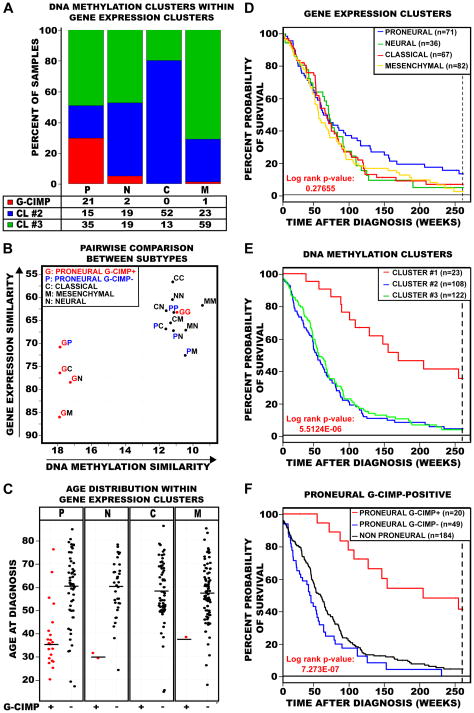
Figure 2. Characterization of G-CIMP tumors as a unique subtype of GBMs within the proneural gene expression subgroup.
A) Integration of the samples within each DNA methylation and gene expression cluster. Samples are primarily categorized by their gene expression subtype: P, proneural; N, neural; C, classical; M, mesenchymal. The number and percent of tumors within each DNA methylation cluster (red, cluster 1 (G-CIMP); blue, cluster 2; green, cluster 3) are indicated for each gene expression subtype. B) Scatter plot of pairwise comparison of the gene expression and DNA methylation clusters as identified in Figure 1 and Figure S1. Same two-letter represents self-comparison while mixed two-letter represents the pair-wise correlation between gene expression and DNA methylation. Axes are reversed to illustrate increasing similarity. C) GBM patient age distribution at time of diagnosis within each gene expression cluster. Samples are divided by gene expression clusters as identified along the top of each jitter plot, and further subdivided by G-CIMP status within each expression subgroup. G-CIMP-positive samples are indicated as red data points and G-CIMP-negative samples are indicated as black data points. Median age at diagnosis is indicated for each subgroup by a horizontal solid black line. D–F) Kaplan-Meier survival curves for GBM methylation and gene expression subtypes. In each plot, the percent probability of survival is plotted versus time since diagnosis in weeks. All samples with survival data greater than five years were censored. D) Kaplan-Meier survival curves among the four GBM expression subtypes. Proneural tumors are in blue, Neural tumors are in green, Classical tumors are in red, and Mesenchymal tumors are in gold. E) Kaplan-Meier survival curves between the three DNA methylation clusters. Cluster 1 tumors are in red, cluster 2 tumors are in blue and cluster 3 tumors are in green. F) Kaplan-Meier survival curves between proneural G-CIMP-positive, proneural G-CIMP-negative and all non-proneural GBM tumors. Proneural G-CIMP-positive tumors are in red, proneural G-CIMP-negative tumors are in blue and all non-proneural GBM tumors are in black. See also Figure S2.
In order to obtain an integrated view of the relationships of G-CIMP status and gene expression differences, we performed pairwise comparisons between members of different molecular subgroups (Figure 2B). We calculated the mean Euclidean distance in both DNA methylation and expression for each possible pairwise combination of the five different subtypes: G-CIMP-positive proneural, G-CIMP-negative proneural, classical, mesenchymal and neural tumors. We observed the high dissimilarity of the GP, GN, GC, and GM pairs (Figure 2B), supporting the hypothesis that G-CIMP-positive tumors are a unique molecular subgroup of GBM tumors, and more specifically that G-CIMP status provides further refinement of the proneural subset. Indeed, among the proneural tumors, the G-CIMP-positive tumors are distinctly dissimilar to the mesenchymal tumors (GM pair), while the G-CIMP-negative proneural tumors are relatively similar to mesenchymal tumors (PM pair). We focused downstream analyses on comparisons between G-CIMP-positive versus G-CIMP-negative tumors within the proneural subset, to avoid misidentifying proneural features as G-CIMP-associated features.
Clinical characterization of G-CIMP tumors
We further characterized G-CIMP tumors by reviewing the available clinical covariates for each patient. Although patients with proneural GBM tumors are slightly younger (median age, 56 years) than all other non-proneural GBM patients (median age, 57.5 years), this was not statistically significant (p=0.07). However, patients with G-CIMP tumors were significantly younger at the time of diagnosis compared to patients diagnosed with non-G-CIMP proneural tumors (median ages of 36 and 59 years, respectively; p<0.0001; Figure 2C).
The overall survival for patients of the proneural subtype was not significantly improved compared to other gene expression subtypes (Figure 2D), but significant survival differences were seen for groups defined by DNA methylation status (Figure 2E, 2F, Figure S2B). We observed significantly better survival for proneural G-CIMP-positive patients (median survival of 150 weeks) than proneural G-CIMP-negative patients (median survival of 42 weeks) or all other non-proneural GBM patients (median survivals of 54 weeks). G-CIMP status remained a significant predictor of improved patient survival (p=0.0165) in Cox multivariate analysis after adjusting for patient age, recurrent vs. non-recurrent tumor status and secondary GBM vs. primary GBM status.
IDH1 sequence alterations in G-CIMP tumors
Nine genes were found to have somatic mutations that were significantly associated with proneural G-CIMP-positive tumors (Figure S3A, Table S2). IDH1 somatic mutations, recently identified primarily in secondary GBM tumors (Balss et al., 2008; Parsons et al., 2008; Yan et al., 2009), were found to be very tightly associated with G-CIMP in our data set (Table 1), with 18 IDH1 mutations primarily observed in 23 (78%) G-CIMP-positive tumors, and 184 G-CIMP-negative tumors were IDH1-wildtype (p< 2.2×10−16). The five discordant cases of G-CIMP-positive, IDH1-wildtype are not significantly different in age compared to G-CIMP-positive, IDH1-mutant (median ages of 34 and 37 years respectively; p=0.873). However, the five discordant cases of G-CIMP-positive, IDH1-wildtype tumors are significantly younger at the time of diagnosis compared to patients with G-CIMP-negative, IDH1-wildtype (median ages of 37 and 59 years respectively; p<0.008). Interestingly, two of the five patients each survived more than five years after diagnosis. We did not observe any IDH2 mutations in the TCGA data set. Tumors displaying both G-CIMP-positive and IDH1 mutation occurred at low frequency in primary GBM, but were enriched in the set of 16 recurrent (treated) tumors, and to an even greater degree in the set of four secondary GBM (Table 1). We also identified examples of germline mutations (nine genes) and loss of heterozygosity (six genes) that were significantly associated with proneural G-CIMP-positive tumors (Figure S3B-C, Table S2).
Table 1. G-CIMP and IDH1 mutation status in primary, secondary, and recurrent GBMs.
G-CIMP and IDH1 mutation status are compared for All analyzed GBM tumors (p< 2.2x10−16), Primary tumors, Recurrent tumors, and Secondary tumors. P value is calculated from Fisher’s exact test. See also Figure S3 and Table S2.
| ALL TUMORS | G-CIMP | TOTAL | ||
|---|---|---|---|---|
| − | + | |||
| IDH1 | Wild-type | 184 | 5 | 189 |
| Mutant | 0 | 18 | 18 | |
| TOTAL | 184 | 23 | 207 | |
| PRIMARY TUMORS | G-CIMP | TOTAL | ||
| − | + | |||
| IDH1 | Wild-type | 171 | 4 | 175 |
| Mutant | 0 | 12 | 12 | |
| TOTAL | 171 | 16 | 187 | |
| RECURRENT TUMORS | G-CIMP | TOTAL | ||
| − | + | |||
| IDH1 | Wild-type | 12 | 0 | 12 |
| Mutant | 0 | 4 | 4 | |
| TOTAL | 12 | 4 | 16 | |
| SECONDARY | G-CIMP | TOTAL | ||
| TUMORS | − | + | ||
| IDH1 | Wild-type | 1 | 1 | 2 |
| Mutant | 0 | 2 | 2 | |
| TOTAL | 1 | 3 | 4 | |
Copy number variation (CNV) in proneural G-CIMP tumors
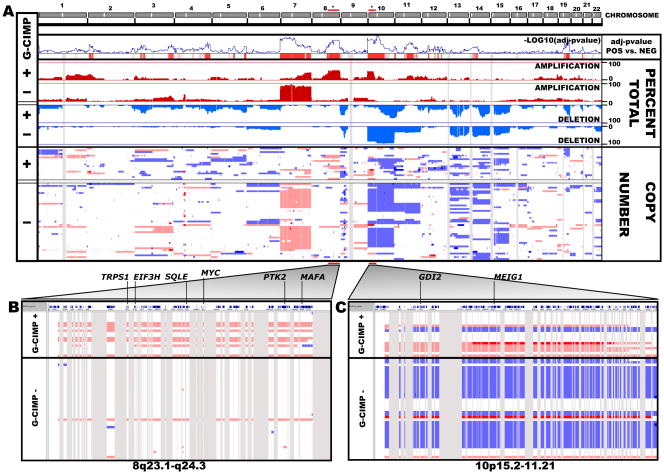
In order to elucidate critical alterations within proneural G-CIMP-positive tumors, we analyzed gene-centric copy number variation data (see Supplemental Information). We identified significant copy number differences in 2,875 genes between proneural G-CIMP-positive and G-CIMP-negative tumors (Figure 3A, Table S3). Although chromosome 7 amplifications are a hallmark of aggressive GBM tumors (The Cancer Genome Atlas Research Network, 2008), copy number variation along chromosome 7 was reduced in proneural G-CIMP-positive tumors. Gains in chromosomes 8q23.1-q24.3 and 10p15.3-p11.21 were identified (Figure 3B and 3C). The 8q24 region contains the MYC oncogene, is rich in sequence variants and was previously shown as a risk factor for several human cancers (Amundadottir et al., 2006; Freedman et al., 2006; Haiman et al., 2007a; Haiman et al., 2007b; Schumacher et al., 2007; Shete et al., 2009; Visakorpi et al., 1995; Yeager et al., 2007). Accompanying the gains at chromosome 10p in proneural G-CIMP-positive tumors, we also detected deletions of the same chromosome arm in G-CIMP-negative tumors (Figure 3C). Similar copy number variation results were obtained when comparing all G-CIMP-positive to G-CIMP-negative samples (Figure S4). These findings point to G-CIMP-positive tumors as having a distinct profile of copy number variation when compared to G-CIMP-negative tumors.
Figure 3. Significant regions of copy number variation in the G-CIMP genome.
Copy number variation for 23,748 loci (across 22 autosomes and plotted in genomic coordinates along the x-axis) was analyzed using 61 proneural TCGA GBM tumors. Homozygous deletion is indicated in dark blue, hemizygous deletion in light blue, neutral/no change in white, gain in light red and high-level amplification in dark red. A) Copy number variation between proneural G-CIMP positive and G-CIMP negative tumors. The Cochran Armitage test for trend, percent total amplification/deletion and raw copy number values are listed. The – log10(FDR-adjusted P value) between G-CIMP-positive and G-CIMP negative proneurals is plotted along the y-axis in the “adjusted P-value pos vs. neg” panel. In this panel, red vertical lines indicate significance. Gene regions in 8q23.1-q24.3 and 10p15.2-11.21 are identified by asterisks and are highlighted in panels B and C, respectively. See also Figure S4 and Table S3.
Identification of DNA methylation and transcriptome expression changes in proneural G-CIMP tumors
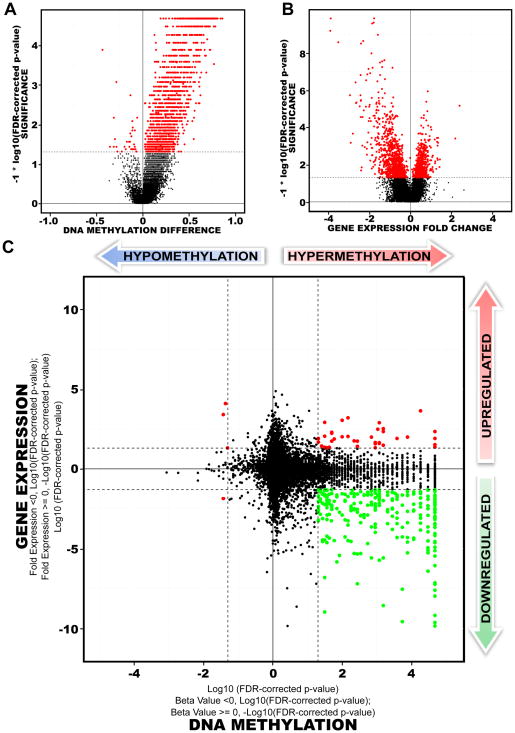
To better understand CpG island hypermethylation in glioblastoma, we investigated the differentially methylated CpG sites of these samples (Figure 1B). Among 3,153 CpG sites that were differentially methylated between proneural G-CIMP positive and proneural G-CIMP-negative tumors, 3,098 (98%) were hypermethylated (Figure 4A). In total, there were 1,550 unique genes, of which 1,520 were hypermethylated and 30 were hypomethylated within their promoter regions. We ranked our probe list by decreasing adjusted p-values and increasing beta-value difference in order to identify the top most differentially hypermethylated CpG probes within proneural G-CIMP-positive tumors (Table S3).
Figure 4. Comparison of transcriptome versus epigenetic differences between proneural G-CIMP-positive and G-CIMP-negative tumors.
A) Volcano Plots of all CpG loci analyzed for G-CIMP association. The beta value difference in DNA Methylation between the proneural G-CIMP-positive and proneural G-CIMP-negative tumors is plotted on the x-axis, and the p-value for a FDR-corrected Wilcoxon signed-rank test of differences between the proneural G-CIMP-positive and proneural G-CIMP-negative tumors (−1* log10 scale) is plotted on the y-axis. Probes that are significantly different between the two subtypes are colored in red. B) Volcano plot for all genes analyzed on the Agilent gene expression platform. C) Starburst plot for comparison of TCGA Infinium DNA methylation and Agilent gene expression data normalized by copy number information for 11,984 unique genes. Log10(FDR-adjusted P value) is plotted for DNA methylation (x-axis) and gene expression (y-axis) for each gene. If a mean DNA methylation β-value or mean gene expression value is higher (greater than zero) in G-CIMP-positive tumors, −1 is multiplied to log10(FDR-adjusted P value), providing positive values. The dashed black lines indicates FDR-adjusted P value at 0.05. Data points in red indicate those that are significantly up- and down-regulated in their gene expression levels and significantly hypo- or hypermethylated in proneural G-CIMP-positive tumors. Data points in green indicate genes that are significantly down-regulated in their gene expression levels and hypermethylated in proneural G-CIMP-positive tumors compared to proneural G-CIMP-negative tumors. See also Figure S5 and Table S3.
The Agilent transcriptome data were used to detect genes showing both differential expression and G-CIMP DNA methylation, in G-CIMP-positive and negative proneural samples. Gene expression values were adjusted for regional copy number changes, as described in Supplemental Information. A total of 1,030 genes were significantly down-regulated and 654 genes were significantly up-regulated among proneural G-CIMP-positive tumors (Figure 4B, Table S3). The differentially down-regulated gene set was highly enriched for polysaccharide, heparin and glycosaminoglycan binding, collagen, thrombospondin and cell morphogenesis (p<2.2E-04, Table S3). In addition, the significantly up-regulated gene set was highly enriched among functional categories involved in regulation of transcription, nucleic acid synthesis, metabolic processes, and cadherin-based cell adhesion (p<6.2E-04). Zinc finger transcription factors were also found to be highly enriched in genes significantly up-regulated in expression (p=3.1E-08). Similar findings were obtained when a permutation analysis was performed and when Affymetrix gene expression data were used (data not shown). We also identified 20 miRNAs that showed significant differences in their gene expression between proneural G-CIMP-positive and proneural G-CIMP-negative tumors (Figure S5, Table S3).
Integration of the normalized gene expression and DNA methylation gene lists identified a total of 300 genes with both significant DNA hypermethylation and gene expression changes in G-CIMP-positive tumors compared to G-CIMP-negative tumors within the proneural subset. Of these, 263 were significantly down-regulated and hypermethylated within proneural G-CIMP-positive tumors (Figure 4C, lower right quadrant). To validate these differentially expressed and methylated genes, we replicated the analysis using an alternate expression platform (Affymetrix), and derived consistent results (Figure S5). Among the top ranked genes were FABP5, PDPN, CHI3L1 and LGALS3 (Table 2), which were identified in an independent analysis to be highly prognostic in GBM with higher expression associated with worse outcome (Colman et al., 2010). Gene ontology analyses showed G-CIMP-specific down-regulation of genes associated with the mesenchyme subtype, tumor invasion and the extracellular matrix as the most significant terms (Table S3). Genes with roles in transcriptional silencing, chromatin structure modifications and activation of cellular metabolic processes showed increased gene expression in proneural G-CIMP-positive tumors. Additional genes differentially expressed in proneural G-CIMP-positive samples are provided in Table S3.
Table 2. The top 50 most differentially hypermethylated and down regulated genes in proneural G-CIMP positive tumors.
Genes are sorted by decreasing Gene Expression log2 ratios. Beta value difference indicates differences in mean of beta values (DNA methylation values) between proneural G-CIMP-positive and proneural G-CIMP-negative. Fold Change is the log2 ratio of the means of proneural G-CIMP-positive and proneural G-CIMP-negative normalized expression intensities. See also Table S3.
| Gene Name | DNA Methylation | Gene Expression | ||
|---|---|---|---|---|
| Wilcoxon Rank P-Value | Beta Value Difference | Wilcoxon Rank P-Value | Fold Change | |
| G0S2 | 2.37E-07 | 0.76 | 2.12E-13 | −3.92 |
| RBP1 | 2.37E-07 | 0.84 | 1.07E-14 | −3.9 |
| FABP5 | 2.30E-05 | 0.3 | 1.39E-12 | −3.53 |
| CA3 | 4.50E-06 | 0.43 | 2.06E-06 | −2.82 |
| RARRES2 | 4.74E-07 | 0.63 | 6.25E-10 | −2.69 |
| OCIAD2 | 2.06E-04 | 0.32 | 1.23E-08 | −2.64 |
| CBR1 | 3.30E-05 | 0.37 | 3.77E-09 | −2.45 |
| PDPN | 3.87E-03 | 0.23 | 1.47E-07 | −2.43 |
| LGALS3 | 2.37E-07 | 0.72 | 7.97E-09 | −2.42 |
| CTHRC1 | 3.50E-04 | 0.45 | 1.44E-07 | −2.34 |
| CCNA1 | 4.66E-03 | 0.29 | 2.95E-05 | −2.14 |
| ARMC3 | 1.76E-03 | 0.31 | 7.76E-05 | −2.13 |
| CHST6 | 5.74E-04 | 0.22 | 8.70E-06 | −2.11 |
| C11orf63 | 2.37E-07 | 0.64 | 7.95E-11 | −2.05 |
| GJB2 | 2.16E-03 | 0.24 | 2.94E-07 | −2.04 |
| KIAA0746 | 1.66E-06 | 0.58 | 2.97E-07 | −1.94 |
| MOSC2 | 2.37E-07 | 0.66 | 6.85E-12 | −1.91 |
| CHI3L1 | 7.11E-06 | 0.13 | 4.11E-06 | −1.9 |
| RARRES1 | 6.38E-05 | 0.41 | 5.93E-09 | −1.89 |
| AQP5 | 4.50E-06 | 0.43 | 6.86E-14 | −1.87 |
| SPON2 | 8.68E-05 | 0.3 | 2.50E-05 | −1.87 |
| RAB36 | 2.06E-04 | 0.26 | 6.49E-11 | −1.86 |
| CHRDL2 | 2.64E-03 | 0.07 | 1.15E-07 | −1.81 |
| TOM1L1 | 2.37E-07 | 0.66 | 4.57E-14 | −1.8 |
| BIRC3 | 2.37E-07 | 0.66 | 6.50E-07 | −1.78 |
| LDHA | 4.50E-04 | 0.55 | 1.83E-07 | −1.74 |
| SEMA3E | 2.37E-07 | 0.52 | 2.36E-04 | −1.72 |
| FMOD | 6.38E-05 | 0.42 | 2.05E-04 | −1.72 |
| C10orf107 | 1.59E-05 | 0.63 | 4.11E-06 | −1.71 |
| FLNC | 2.37E-07 | 0.74 | 1.36E-05 | −1.67 |
| TMEM22 | 4.74E-07 | 0.59 | 1.63E-08 | −1.67 |
| TCTEX1D1 | 1.66E-06 | 0.37 | 2.97E-07 | −1.67 |
| DKFZP586H2123 | 1.59E-05 | 0.53 | 4.83E-05 | −1.66 |
| TRIP4 | 4.74E-07 | 0.49 | 4.18E-08 | −1.65 |
| SLC39A12 | 1.81E-03 | 0.2 | 7.24E-06 | −1.65 |
| FLJ21963 | 4.60E-05 | 0.26 | 8.07E-07 | −1.63 |
| CRYGD | 4.74E-07 | 0.54 | 2.81E-08 | −1.62 |
| LECT1 | 5.74E-04 | 0.32 | 3.73E-07 | −1.61 |
| EPHX2 | 2.37E-07 | 0.53 | 3.73E-06 | −1.6 |
| LGALS8 | 3.20E-03 | 0.19 | 4.15E-13 | −1.6 |
| C7orf46 | 2.37E-07 | 0.35 | 4.11E-06 | −1.58 |
| F3 | 2.37E-07 | 0.62 | 4.76E-08 | −1.57 |
| TTC12 | 9.48E-07 | 0.53 | 4.52E-06 | −1.57 |
| ITGBL1 | 1.42E-03 | 0.2 | 1.86E-07 | −1.57 |
| B3GNT5 | 1.66E-06 | 0.66 | 2.36E-07 | −1.55 |
| NMNAT3 | 2.30E-05 | 0.62 | 5.93E-09 | −1.55 |
| FZD6 | 6.38E-05 | 0.34 | 2.28E-06 | −1.55 |
| FKBP5 | 9.16E-04 | 0.23 | 9.22E-09 | −1.54 |
| SLC25A20 | 1.56E-04 | 0.6 | 1.41E-08 | −1.53 |
| MMP9 | 2.06E-04 | 0.37 | 2.09E-03 | −1.53 |
To extend these findings, the differentially silenced genes were subjected to a NextBio (www.nextbio.com) meta-analysis to identify data sets that were significantly associated with our list of 263 hypermethylated and down-regulated genes. There was an overlap with down-regulated genes in low- and intermediate-grade glioma compared to GBM in a variety of previously published datasets (Ducray et al., 2008; Liang et al., 2005; Sun et al., 2006) (Figure S5, Table S3). The overlap of the 263-gene set with each of these additional datasets was unlikely to be due to chance (all analyses p<0.00001). To further characterize this gene set, we tested the survival association of these gene expression values in a collection of Affymetrix profiling data from published and publicly available sources on which clinical annotation was available. This dataset included cohorts from the Rembrandt set (Madhavan et al., 2009) as well as other sources, and did not include TCGA data (since TCGA data were used to derive the gene list). In this combined dataset, the expression of the 263-gene set was significantly associated with patient outcome (Figure S5G). Together, these findings suggest that G-CIMP-positive GBMs tumors have epigenetically-related gene expression differences which are more consistent with low-grade gliomas as well as high grade tumors with favorable prognosis.
Validation of G-CIMP in GBM and incidence in low-grade gliomas
To validate the existence of G-CIMP loci and better characterize the frequency of G-CIMP in gliomas, MethyLight was used to assay the DNA methylation levels in eight G-CIMP gene regions in seven hypermethylated loci (ANKRD43, HFE, MAL, LGALS3, FAS-1, FAS-2, and RHO-F) and one hypomethlyated locus, DOCK5, in the tumor samples. These eight markers were evaluated in paraffin embedded tissues from 20 TCGA samples of known G-CIMP status (10 G-CIMP-postive and 10 G-CIMP negative). We observed perfect concordance between G-CIMP calls on the array platforms versus with the MethyLight markers, providing validation of the technical performance of the platforms and of the diagnostic marker panel. These 20 samples were excluded from the validation set described below. A sample was considered G-CIMP positive if at least six genes displayed a combination of DOCK5 DNA hypomethylation and/or hypermethylation of the remaining genes in the panel. Using these criteria, we tested an independent set of non-TCGA GBM samples for G-CIMP status. Sixteen of 208 tumors (7.6%) were found to be G-CIMP-positive (Figure 5A), very similar to the findings in TCGA data.
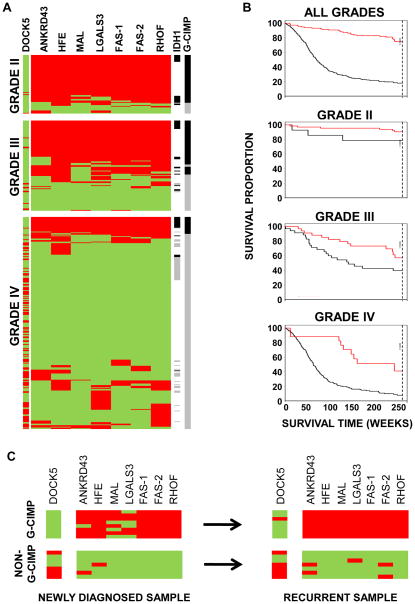
Figure 5. G-CIMP prevalence in grade II, III and IV gliomas using MethyLight.
A) Methylation profiling of gliomas shows an association of CIMP status with tumor grade. Eight markers were tested for G-CIMP DNA methylation in 360 tumor samples. Each marker was coded as red if methylated and green if unmethylated. One of these markers (DOCK5) is unmethylated in CIMP, while the remaining seven markers show G-CIMP-specific hypermethylation. G-CIMP-positive status was determined if ≥6 of the 8 genes had G-CIMP-defining hyper- or hypomethylation. G-CIMP-positive status is indicated with a black line (right side of panel), and a grey line indicates non G-CIMP. Samples with an identified IDH1 mutation is indicated as a black line and samples with no known IDH1 mutation as a grey line. White line indicates unknown IDH1 status. B) Association of G-CIMP status with patient outcome stratified by tumor grade. G-CIMP-positive cases are indicated by red lines and the G-CIMP-negative cases are indicated by black lines in each Kaplan-Meier survival curve. C) Stability of G-CIMP over time in glioma patients. Fifteen samples from newly diagnosed tumors were tested for G-CIMP positivity using the eight-marker MethyLight panel. Eight tumors were classified as G-CIMP-positive (upper left panel), and seven tumors were classified as G-CIMP-negative (non-G-CIMP, lower left panel). Samples from a second procedure, ranging from 2–9 years after the initial resection, were also evaluated for the G-CIMP-positive cases (upper right panel), as well as for the non-G-CIMP cases (lower right panel). Each marker was coded as red if methylated and green if unmethylated.
To further expand these observations, we determined the IDH1 mutation status for an independent set 100 gliomas (WHO grades II, III and IV). Among 48 IDH1-mutant tumors, 35 (72.9%) were G-CIMP. However, only 3/52 cases (5.8%) without an IDH1 mutation were G-CIMP positive (odds ratio= 42; 95% confidence interval (CI), 11–244; Figure S3D), validating the tight association of G-CIMP and IDH1 mutation.
Based on the association of G-CIMP status with features of the progressive, rather than the de novo GBM pathway, we hypothesized that G-CIMP status was more common in the low- and intermediate-grade gliomas. We extended this analysis by evaluating 60 grade II and 92 grade III gliomas for G-CIMP DNA methylation using the eight gene MethyLight panel. Compared to GBM, grade II tumors showed an approximately 10-fold increase in G-CIMP-positive tumors, while grade III tumors had an intermediate proportion of tumors that were G-CIMP-positive (Figure 5A, Figure S3E). When low- and intermediate-grade gliomas were separated by histologic type, G-CIMP positivity appeared to be approximately twice as common in oligodendrogliomas (52/56, 93%) as compared to astrocytomas (43/95, 45%). G-CIMP positive status correlated with improved patient survival within each WHO-recognized grade of diffuse glioma, indicating that the G-CIMP status was prognostic for glioma patient survival (p<0.032, Figure 5B). G-CIMP status was an independent predictor (p<0.01) of survival after adjustment for patient age and tumor grade (Figure S3F). Together, these findings show that G-CIMP is a prevalent molecular signature in low grade gliomas and confers improved survival in these tumors.
Stability of G-CIMP at recurrence
Since epigenetic events can be dynamic processes, we examined whether G-CIMP status was a stable event in glioma or whether it was subject to change over the course of the disease. To test this, we obtained a set of samples from 15 patients who received a second surgical procedure following tumor recurrence, with time intervals of up to eight years between initial and second surgical procedures. We used the eight-gene MethyLight panel to determine their G-CIMP status and found that eight samples were G-CIMP-positive, while seven were G-CIMP-negative. Interestingly, among the G-CIMP-positive cases, 8/8 (100%) recurrent samples retained their G-CIMP positive status. Similarly, among seven G-CIMP-negative cases, all seven remained G-CIMP-negative at recurrence, indicating stability of the G-CIMP phenotype over time (Figure 5C).
Discussion
In this report, we identified and characterized a distinct molecular subgroup in human gliomas. Analysis of epigenetic changes from TCGA samples identified the existence of a proportion of GBM tumors with highly concordant DNA methylation of a subset of loci, indicative of a CpG island methylator phenotype (G-CIMP). G-CIMP-positive samples were associated with secondary or recurrent (treated) tumors and tightly associated with IDH1 mutation. G-CIMP tumors also showed a relative lack of copy number variation commonly observed in GBM, including EGFR amplification, chromosome 7 gain and chromosome 10 loss. Interestingly, G-CIMP tumors displayed copy-number alterations which were also shown in gliomas with IDH1 mutations in a recent report (Sanson et al., 2009). Integration of the DNA methylation data with gene expression data showed that G-CIMP-positive tumors represent a subset of proneural tumors. G-CIMP-positive tumors showed a favorable prognosis within GBMs as a whole and also within the proneural subset, consistent with prior reports for IDH1 mutant tumors (Parsons et al., 2008; Yan et al., 2009). Interestingly, of the five discordant cases of G-CIMP-positive, IDH1-wildtype tumors, two patients survived more than five years after diagnosis, suggesting that G-CIMP-positive status may confer favorable outcome independent of IDH1 mutation status. However, studies with many more discordant cases will be needed to carefully dissect the effects of G-CIMP status versus IDH1 mutation on survival. To a large extent, the improved prognosis conferred by proneural tumors (Phillips et al., 2006) can be accounted for by the G-CIMP-positive subset. These findings indicate that G-CIMP could be use to further refine the expression-defined groups into an additional subtype with clinical implications.
G-CIMP is highly associated with IDH1 mutation across all glioma tumor grades, and the prevalence of both decreases with increasing tumor grade. Tumor grade is defined by morphology only, and therefore can be heterogeneous with respect to molecular subtypes. Within grade IV/glioblastoma tumors are a subset of patients who tend to be younger and have a relatively favorable prognosis. It is only through molecular characterization using markers such as IDH1 and G-CIMP status that one could prospectively identify such patients. Conversely, these markers could also be used to identify patients with low- and intermediate-grade gliomas who may exhibit unfavorable outcome relative to tumor grade.
In the non-TCGA independent validation set examined in this study, an IDH1 mutation was detected in 40/43 (93%) low- and intermediate-grade gliomas, but only 7/57 (13%) of primary GBMs. Similarly, we detected nearly 10-fold more G-CIMP-positive gliomas in grade II tumors as compared to grade IV GBMs. The improved survival of G-CIMP gliomas at all tumor grades suggests that there are molecular features within G-CIMP gliomas that encourage a less aggressive tumor phenotype. Consistent with this, we identified G-CIMP-specific DNA methylation changes within a broad panel of genes whose expression was significantly associated with patient outcome. We observed that this large subset of differentially silenced genes were involved in specific functional categories, including markers of mesenchyme, tumor invasion and extracellular matrix. This concept builds upon our prior finding of a mesenchymal subgroup of glioma which shows poor prognosis (Phillips et al., 2006). According to this model, a lack of methylation of these genes in G-CIMP-negative tumors would result in a relative increase in expression of these genes, which in turn would promote tumor progression and/or lack of response to currently available treatment modalities. A comparison of the G-CIMP gene list with prior gene expression analyses (meta-analyses) suggests that G-CIMP positive tumors may be less aggressive due to silencing of key mesenchymal genes.
We found that a minority of genes with significant promoter hypermethylation showed a concomitant significant decrease in associated gene expression (293/1520, 19%). This is consistent with previous reports, in which we found similarly low frequencies of inversely correlated promoter hypermethylation and gene expression (Houshdaran et al., 2010; Pike et al., 2008). The lack of an inverse relationship between promoter hypermethylation and gene expression for most genes may be attributed to several scenarios, including the lack of appropriate transcription factors for some unmethylated genes and the use of alternative promoters for some genes with methylated promoters. Epigenetics controls expression potential, rather than expression state.
RBP1 and G0S2 are the two genes showing the strongest evidence for epigenetic silencing in G-CIMP tumors. RBP1 has been previously reported to be epigenetically silenced in cancer cell lines and primary tumors, and the association of its encoded protein with retinoic acid receptors (RAR) has been well characterized (Esteller et al., 2002). G0S2 gene expression is regulated by retinoic acid (RA), and encodes a protein that promotes apoptosis in primary cells suggesting a tumor suppressor role (Kitareewan et al., 2008; Welch et al., 2009). The vitamin A metabolite RA is important for both embryonic and adult growth. RA has a diverse roles involving neuronal development and differentiation mediated by RARs (reviewed in (Malik et al., 2000)). Interestingly, studies in breast cancer cells have shown that silencing of RBP1 plays an important role in RA signaling by lowering all-trans-retinoic acid production and loss of RAR levels and activity mediated by de-repression of PI3K/Akt signaling pathway, leading to loss of cell differentiation and tumor progression ((Farias et al., 2005a),(Farias et al., 2005b)). This mechanism may help describe the molecular features of tumorigenesis in G-CIMP tumors. Thus, dissecting the gene expression and DNA methylation alterations of G-CIMP tumors among lower grade gliomas will be helpful to better understand the roles of a mutant IDH1 and G-CIMP DNA methylation on tumor grade and patient survival.
The highly concerted nature of G-CIMP methylation suggests that this phenomenon may be caused by a defect in a trans-acting factor normally involved in the protection of a defined subset of CpG island promoters from encroaching DNA methylation. Loss of function of this factor would result in widespread concerted DNA methylation changes. We propose that transcriptional silencing of some CIMP genes may provide a favorable context for the acquisition of specific genetic lesions. Indeed, we have recently found that IGFBP7 is silenced by promoter hypermethylation in BRAF-mutant CIMP+ colorectal tumors (Hinoue et al., 2009). Oncogene-induced senescence by mutant BRAF is known to be mediated by IGFBP7 (Wajapeyee et al., 2008). Hence, CIMP-mediated inactivation of IGFBP7 provides a suitable environment for the acquisition of BRAF mutation. The tight concordance of G-CIMP status with IDH1 mutation in GBM tumors is very reminiscent of colorectal CIMP, in which DNA hypermethylation is strongly associated with BRAF mutation (Weisenberger et al., 2006). We hypothesize that the transcriptional silencing of as yet unknown G-CIMP targets may provide an advantageous environment for the acquisition of IDH1 mutation.
In our integrative analysis of G-CIMP tumors, we observed up-regulation of genes functionally related to cellular metabolic processes and positive regulation of macromolecules. This expression profile may reflect a metabolic adjustment to the proliferative state of the tumor, in conjunction with the gain-of-function IDH1 mutation (Dang et al., 2009). Such a metabolic adjustment may be consistent with Warburg’s observation that proliferating normal and tumor cells require both biomass and energy production, and convert glucose primarily to lactate, regardless of oxygen levels, while non-proliferating differentiated cells emphasize efficient energy production (reviewed in (Vander Heiden et al., 2009)).
In summary, our data indicate that G-CIMP status stratify gliomas into two distinct subgroups with different molecular and clinical phenotypes. These molecular classifications have implications for differential therapeutic strategies for glioma patients. Further observation and characterization of molecular subsets of glioma will likely provide additional information enabling insights into the the development and progression of glioma, and may lead to targeted drug treatment for patients with these tumors.
Experimental Procedures
Samples and DNA methylation assays
Genomic DNAs from TCGA GBM tumors were isolated by the TCGA Biospecimen Core Resource (BCR) and delivered to USC as previously described (The Cancer Genome Atlas Research Network, 2008). One sample (TCGA-06-0178) with a confirmed IDH1 mutation was removed from our analyses, since it became clear that an incorrect tissue type had been shipped for the DNA methylation analysis. Four brain genomic DNA samples from apparently healthy individuals were included as controls. All tissue samples (patients and healthy individuals) were obtained using institutional review board–approved protocols from University of Southern California (TCGA GBM samples), Johns Hopkins School of Medicine (Normal Brain samples) and The University of Texas MD Anderson Cancer Center (Glioma validation samples). Tissue samples were deidentified to ensure patient confidentiality. Genomic DNA methylated in vitro with M.SssI methylase or whole genome amplified (WGA) as positive and negative controls for DNA methylation, respectively, were also included. Details are in Supplemental Information.
The GoldenGate assays were performed according to the manufacturer and as described previously (Bibikova et al., 2006). The GoldenGate methylation assays survey the DNA methylation of up to 1,536 CpG sites - a total of 1,505 CpGs spanning 807 unique gene loci are interrogated in the OMA-002 probe set (Bibikova et al., 2006), and 1,498 CpGs spanning the same number of unique gene regions are investigated in the OMA-003 probe set (The Cancer Genome Atlas Research Network, 2008).
The Infinium methylation assays were performed according to manufacturer’s instructions. The assay generates DNA methylation data for 27,578 CpG dinucleotides spanning 14,473 well-annotated, unique gene promoter and/or 5′ gene regions (from −1,500 to +1,500 from the transcription start site). The assay information is available at www.illumina.com and the probe information is available on the TCGA Data Portal web site. Data from 91 TCGA GBM samples (Batches 1, 2, 3 and 10) were included in this analysis. Batches 1–3 (63 samples) were run on both Inifium and GoldenGate, whereas batches 4–8 (182 samples) were analyzed exclusively on GoldenGate and batch 10 (28 samples) were analyzed exclusively on Infinium. All data were packaged and deposited onto the TCGA Data Portal web site (http://tcga.cancer.gov/dataportal). Figure S1C illustrates a Venn diagram with the overlapping samples between different DNA methylation platforms. Additional details on DNA methylation detection protocols and TCGA GBM data archived versions are in Supplemental Information.
Integrative TCGA data platforms
While ancillary data (expression, mutation, copy number) were available for additional tumor samples, we only included those samples for which there were DNA methylation profiling (either GoldenGate or Infinium). Since the Agilent gene expression platform contained a greater number of genes for which DNA methylation data were available, we limited our primary analysis to only the Agilent gene expression data set. Where appropriate, we confirmed results using the Affymetrix gene expression data. Additional details are in Supplemental Information.
Clustering analysis and measurement of differential DNA methylation and differential gene expression
Probes for each platform were filtered by removing those targeting the X and Y chromosomes, those containing a single nucleotide polymorphism (SNP) within five basepairs of the targeted CpG site, and probes containing repeat element sequences ≥10 basepairs. We next retained the most variably methylated probes (standard deviation > 0.20) across the tumor set in each DNA methylation platform. These final data matrices were used for unsupervised Consensus/Hierarchical clustering analyses. A non-parametric approach (Wilcoxon rank-sum test) was used to determine probes/genes that are differentially DNA methylated or differentially expressed between the two groups of interest. Additional information is described in Supplemental Information.
G-CIMP validation using MethyLight technology
Tumor samples were reviewed by a neuropathologist (K.A.) to ensure accuracy of diagnosis as well as quality control to minimize normal tissue contamination. MethyLight real-time PCR strategy was performed as described previously (Eads et al., 2000; Eads et al., 1999). Additional details are in Supplemental Information.
Pathway and meta-analyses and statistical analyses
Additional tools included the Molecular Signatures Database (MSigDB database v2.5), Database for Annotation, Visualization and Integrated Discovery (DAVID) and NextBio™. All statistical tests were done using R software (R version 2.9.2, 2009-08-24, (R Development Core Team, 2009)) and packages in Bioconductor (Gentleman et al., 2004), except as noted. Additional details are in Supplemental Information.
Significance
Glioblastoma (GBM) is a highly aggressive form of brain tumor, with a patient median survival of just over one year. The Cancer Genome Atlas (TCGA) project aims to characterize cancer genomes to identify means of improving cancer prevention, detection, and therapy. Using TCGA data, we identified a subset of GBM tumors with characteristic promoter DNA methylation alterations, referred to as a glioma CpG Island Methylator Phenotype (G-CIMP). G-CIMP tumors have distinct molecular features, including a high frequency of IDH1 mutation and characteristic copy-number alterations. Patients with G-CIMP tumors are younger at diagnosis and display improved survival times. The molecular alterations in G-CIMP tumors define a distinct subset of human gliomas with specific clinical features.
Highlights
Identification of a CpG island methylator phenotype (G-CIMP) in gliomas
G-CIMP is tightly associated with IDH1 mutation
G-CIMP patients are younger at diagnosis and display improved survival
G-CIMP is more prevalent among low- and intermediate-grade gliomas
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grants U24 CA126561 and U24 CA143882-01 (P.W.L and S.B.B.) and grants from the Brain Tumor Funders’ Collaborative, the V Foundation, the Rose Foundation and SPORE grant P50CA127001 from NIH/NCI (K.A.). We thank Dennis Maglinte and members of the USC Epigenome Center for helpful discussions, Andreana Rivera for technical assistance and Marisol Guerrero for editorial assistance.
Footnotes
Conflict of interest statement
P.W.L. is a shareholder, consultant and scientific advisory board member of Epigenomics, AG, which has a commercial interest in DNA methylation markers. This work was not supported by Epigenomics, AG. K.A. is a consultant and scientific advisory board member for Castle Biosciences, which has a commercial interest in molecular diagnostics. This work was not supported by Castle Biosciences.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- Brandes AA, Ermani M, Basso U, Amista P, Berti F, Scienza R, Rotilio A, Pinna G, Gardiman M, Monfardini S. Temozolomide as a second-line systemic regimen in recurrent high-grade glioma: a phase II study. Ann Oncol. 2001;12:255–257. doi: 10.1023/a:1008336732273. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoll P, Goldman JE. The interface between glial progenitors and gliomas. Acta Neuropathol. 2008;116:465–477. doi: 10.1007/s00401-008-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. A multigene predictor of outcome in glioblastoma. Neuro-Oncology. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks PB. Cancer: stem cells and brain tumours. Nature. 2006;444:687–688. doi: 10.1038/444687a. [DOI] [PubMed] [Google Scholar]
- Ducray F, Idbaih A, de Reynies A, Bieche I, Thillet J, Mokhtari K, Lair S, Marie Y, Paris S, Vidaud M, et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG. Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res. 2002;62:5902–5905. [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Farias EF, Marzan C, Mira-y-Lopez R. Cellular retinol-binding protein-I inhibits PI3K/Akt signaling through a retinoic acid receptor-dependent mechanism that regulates p85-p110 heterodimerization. Oncogene. 2005a;24:1598–1606. doi: 10.1038/sj.onc.1208347. [DOI] [PubMed] [Google Scholar]
- Farias EF, Ong DE, Ghyselinck NB, Nakajo S, Kuppumbatti YS, Mira y Lopez R. Cellular retinol-binding protein I, a regulator of breast epithelial retinoic acid receptor activity, cell differentiation, and tumorigenicity. J Natl Cancer Inst. 2005b;97:21–29. doi: 10.1093/jnci/dji004. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007a;39:954–56. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007b;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the Association Between CIMP and BRAFV600E in Colorectal Cancer by DNA Methylation Profiling. PLoS ONE. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshdaran S, Hawley S, Palmer C, Campan M, Olsen M, Ventura A, Knudsen B, Drescher C, Urban N, Brown P, Laird PW. DNA methylation profiles of ovarian epithelial carcinoma tumors and cell lines. PLoS ONE. 2010;5:e9359. doi: 10.1371/journal.pone.0009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- Kitareewan S, Blumen S, Sekula D, Bissonnette RP, Lamph WW, Cui Q, Gallagher R, Dmitrovsky E. G0S2 is an all-trans-retinoic acid target gene. Int J Oncol. 2008;33:397–404. [PMC free article] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik MA, Blusztajn JK, Greenwood CE. Nutrients as trophic factors in neurons and the central nervous system: role of retinoic acid. J Nutr Biochem. 2000;11:2–13. doi: 10.1016/s0955-2863(99)00066-2. [DOI] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- Martinez R, Rohde V, Schackert G. Different molecular patterns in glioblastoma multiforme subtypes upon recurrence. J Neurooncol. 2010;96:321–329. doi: 10.1007/s11060-009-9967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Schackert G, Esteller M. Hypermethylation of the proapoptotic gene TMS1/ASC: prognostic importance in glioblastoma multiforme. J Neurooncol. 2007;82:133–139. doi: 10.1007/s11060-006-9264-4. [DOI] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–197. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pike BL, Greiner TC, Wang X, Weisenburger DD, Hsu YH, Renaud G, Wolfsberg TG, Kim M, Weisenberger DJ, Siegmund KD, et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- Schumacher FR, Feigelson HS, Cox DG, Haiman CA, Albanes D, Buring J, Calle EE, Chanock SJ, Colditz GA, Diver WR, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-deficient phenotype in human gliomas: frequency and time to tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–814. [PubMed] [Google Scholar]
- Stone AR, Bobo W, Brat DJ, Devi NS, Van Meir EG, Vertino PM. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am J Pathol. 2004;165:1151–1161. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Tepel M, Roerig P, Wolter M, Gutmann DH, Perry A, Reifenberger G, Riemenschneider MJ. Frequent promoter hypermethylation and transcriptional downregulation of the NDRG2 gene at 14q11.2 in primary glioblastoma. Int J Cancer. 2008;123:2080–2086. doi: 10.1002/ijc.23705. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann K, Rohde K, Zeller C, Szymas J, Vogel S, Marczinek K, Thiel G, Nurnberg P, Laird PW. Distinct methylation profiles of glioma subtypes. Int J Cancer. 2003;106:52–59. doi: 10.1002/ijc.11175. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, TG, Mesirov JP, et al. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Welch C, Santra MK, El-Assaad W, Zhu X, Huber WE, Keys RA, Teodoro JG, Green MR. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782–6789. doi: 10.1158/0008-5472.CAN-09-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.