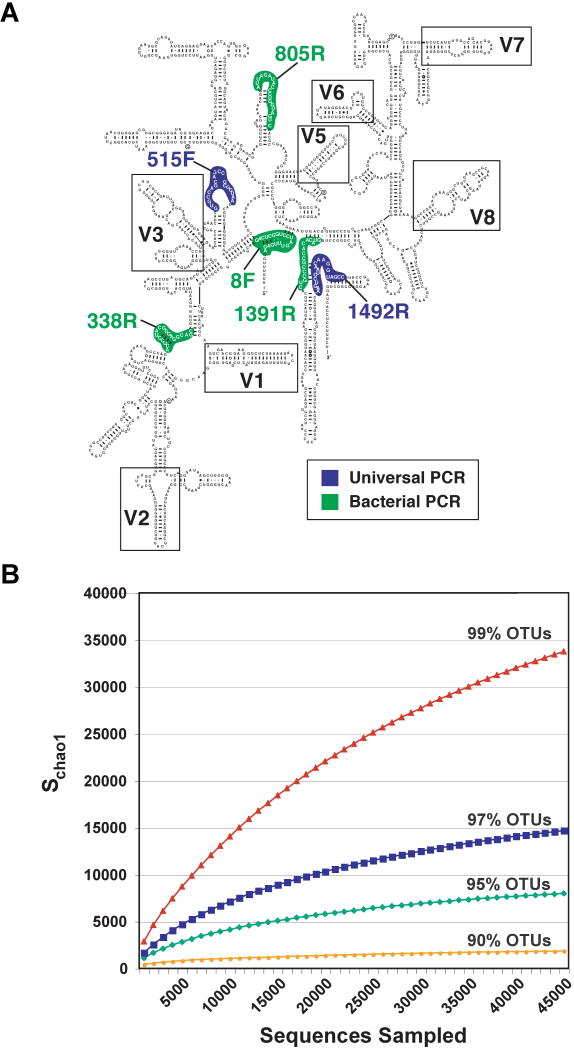
Figure 1. Bacterial SSU rRNA gene-based surveys of the gut microbiota.
(A) Cartoon of the general structure of the bacterial 16S rRNA gene, showing conserved and variable regions (based on http://www.rna.ccbb.utexas.edu/). Targets for PCR primers are shown. The numbers associated with the primers are referenced to the E. coli 16S rDNA gene. The targeted sites for PCR vary depending upon whether the goal is to sequence the entire gene or subdomains containing variable regions. The read length achievable with the current generation of massively parallel sequencers limit the portion of the gene that can be characterized: currently favored regions are V2-V3 (Hamady et al., 2008) and V6 (Huber et al., 2007). (B) Analysis of diversity in the human gut microbial community. Collector's curves of observed and estimated richness are shown. Richness is estimated to be 18,000 (genus-level OTUs with ≥95 %ID) and 36,000 at the species-level (≥99 %ID) using the Chao1 estimator.