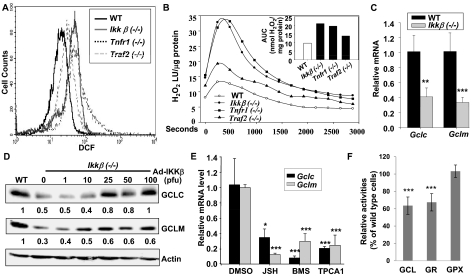
Fig. 3.
Inactivation of the IKKβ pathway reduces GCLC and GCLM expression. Wild-type, Ikkβ(−/−), Tnfr1(−/−), and Traf2(−/−) cells were labeled with CM-H2DCFDA under normal growth conditions and were analyzed by Flow cytometry (A), and used for measurement of H2O2 release by catalase-inhibited luminol chemiluminescence (B). The time-courses for H2O2 release are shown in the line plots as luminescence units (LU)/μg cell protein, with the insert depicting the area under each curve (AUC) as nanomoles of H2O2 release per milligram of protein. Each value represents the average of two experiments. Total RNA was isolated from wild-type and Ikkβ(−/−) cells (C), and wild-type cells were treated for 24 h with 0.1% DMSO, 10 μM JSH23, 1 μM BMS-345541, and 0.5 μM TPCA-1 (E). The RNA were subjected to reverse transcription followed by quantitative RT-PCR. Results showed the levels of Gclc/Gapdh and Gclm/Gapdh in Ikkβ(−/−) relative to those in wild-type cells designated as 1. D, total cell lysates from wild-type, Ikkβ(−/−), and Ikkβ(−/−) cells infected with IKKβ adenovirus were subjected to Western blotting. The GCLC/β-actin and GCLM/β-actin levels were compared with those in wild-type cells designated as 1. F, enzyme activities in Ikkβ(−/−) cells were compared with those in wild-type cells, designated as 100%. All results are presented as the mean values ± S.E. from at least three independent experiments. Statistical analyses were done compared with the mean values in control wild-type cells. **, p < 0.01; ***, p < 0.001 were considered significant.