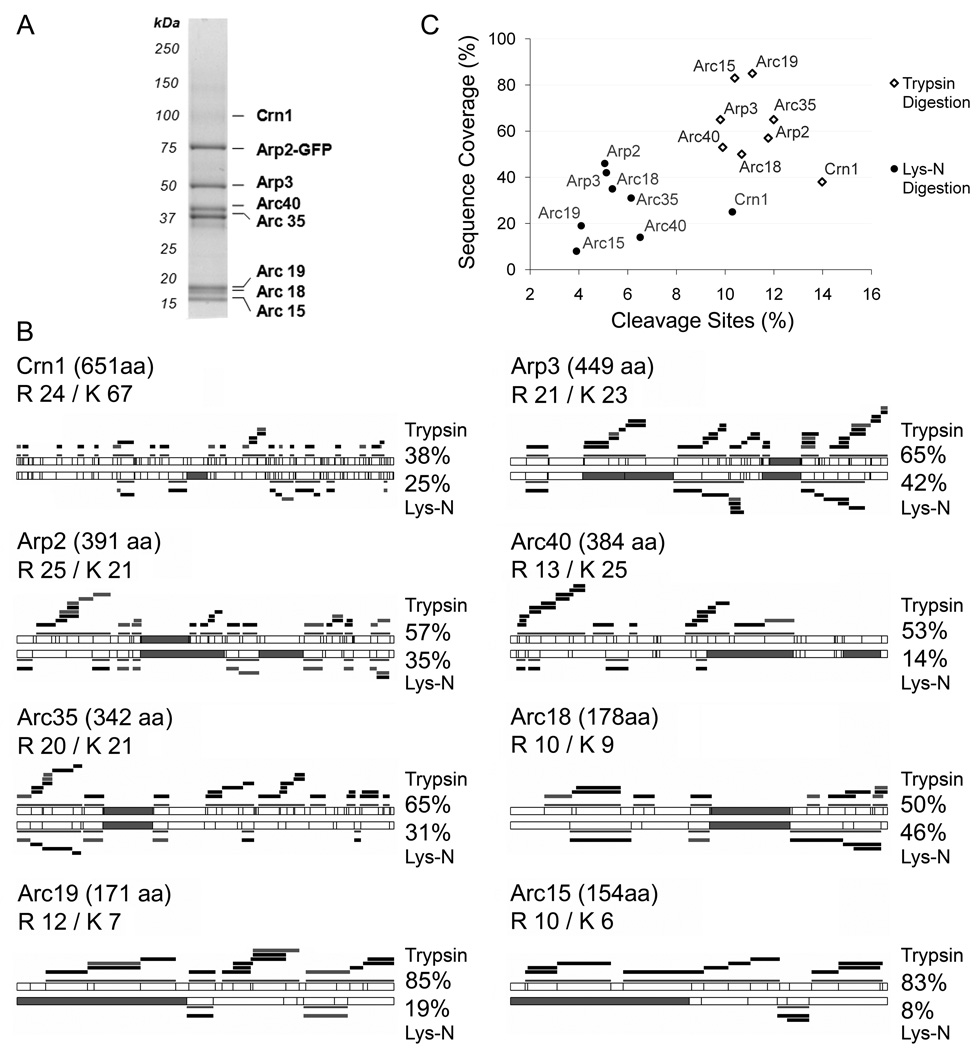
Figure 1. Comparison of protein sequence coverage following trypsin and Lys-N digestion.
(A) A representative SDS-PAGE gel from the immunopurification of Arp2-GFP from yeast. (B) The protein sequence coverages obtained following trypsin (top) and Lys-N (bottom) digestions are represented for the eight proteins from the isolated Arp2 complex. The total numbers of amino acids, R and K residues are indicated. The cleavage sites for trypsin and Lys-N are represented by vertical lines. Filled gray segments represent peptides with an m/z > 4000 that would not be detected by our MALDI LTQ Orbitrap instrument. Detected peptides are represented as black bars for unmodified peptides, and gray bars for peptides that are modified by oxidation of methionine. Projections of the observed peptides and corresponding sequence coverages are indicated. (C) Protein sequence coverage is plotted against the percentage of cleavage sites for the Arp2-isolated proteins following trypsin (open diamonds) and Lys-N (filled circles) digestions.