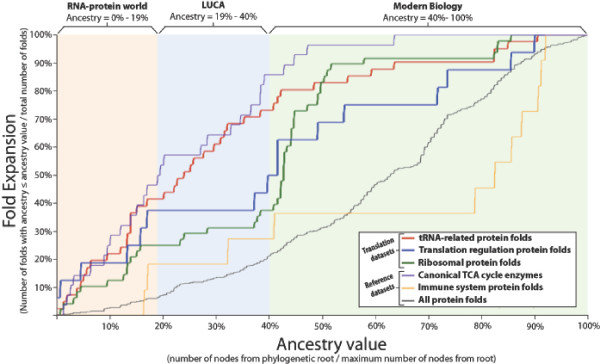
Figure 2.
Protein fold expansion plotted as a function of ancestry. Fold expansion is calculated as the cumulative fraction of folds less than or equal to a given ancestry value. Ancestry values for fold architectures were derived from the phylogenetic tree of all folds by Wang et al. [26] and are equal to the number of nodes from a given fold to the root of the phylogenetic tree divided by the number of nodes from the most recent fold to the root of the tree. Fold expansion can be considered a proxy for sophistication while ancestry value can be considered a proxy for evolutionary time. For reference, the same analysis is performed on canonical TCA cycle enzymes, immune system proteins, and the whole proteome (see Results and discussion). The first fold of a ribonucleotide reductase catalytic domain appears at 19% ancestry, while the first fold found in only one taxonomic domain of life appears at 40% ancestry. We use these values to approximate ranges in ancestry value that correspond to the RNA-protein world, the era of the Last Universal Common Ancestor (LUCA), and the era of modern biology. These results reveal a rapid expansion of translation protein architectures before the divergence of LUCA and even before the establishment of the DNA genome. Quantitative features of these results are presented in Table 1.