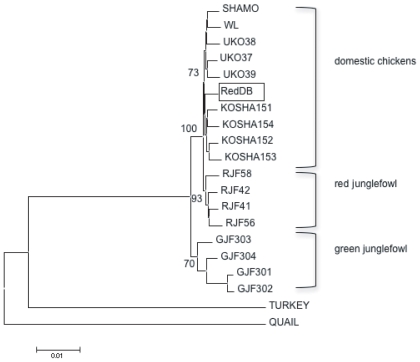
Figure 1. The average-difference tree based on 1,000 concatenated sequences of randomly selected diploid sequences.
The proportion supporting a cluster is shown at each node as the realization of that cluster in the 1,000 individual trees. The TURKEY and QUAIL sequences are used as outgroups. The boxed RedDB indicates that the sequences were taken from the database of red junglefowl. Significant contributions of the domestic chicken genome to this database sequence are evident. The scale shown below the figure is a branch length corresponding to a per-site number of substitutions of 0.01 (1%). Abbreviations for samples are as follows. SHAMO: Shamo, WL: White leghorn, UKO: ukokkei, KOSHA: Koshamo, RJF: red junglefowl, GJF: green junglefowl.