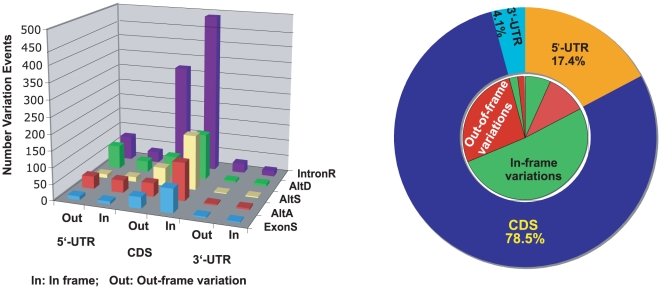
Figure 3. Transcript structure variation (TSV) locations and length difference in Anopheles gambiae.
Left panel shows the detail of variation locations and length differences in different TSV types. For all TSV types, the open reading frames of variations within CDS regions are conserved. Right panel shows the summary of variation locations and length differences. The green and red colors in the inner circle represent in-frame and out-of-frame variations respectively. More than 65% of variations at CDS regions are in-frame, while variations at un-translated regions (UTR) tend to be out-of-frame. Therefore, most transcript structure variations just insert or delete some amino acids without changing the protein structures. The data also show that more variations are at 5′-UTR than at 3′-UTR, which suggests the higher efficiency on gene regulation at 5′-UTR than 3′-UTR.