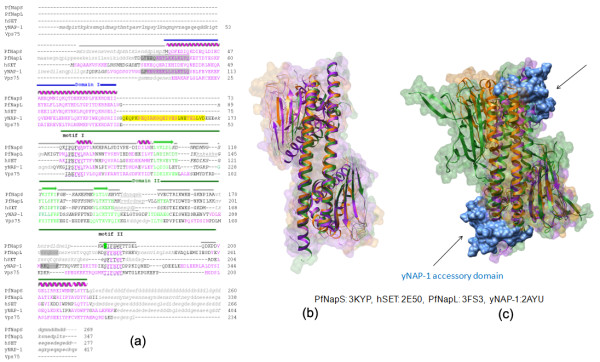
Figure 2.
Comparison of PfNapS with yNAP-1, Vps75, hSET and PfNapL (a). Structure-based sequence alignment of PfNapS, yNAP-1, Vps75, hSET and PfNapL. Residues constituting α-helices and β-sheets in all the individual structures are coloured magenta and green respectively. Domains I and II of PfNapS are indicated using blue and green bars respectively. Conserved hydrophobic motifs are dotted underlined in purple and indicated as 'motif I' and 'motif II'. The 'accessory domain' of yNAP-1 is foreground coloured yellow. The predicted NES and NLS motifs in yNAP-1 are shown in a shaded box coloured grey. The corresponding NES and NLS motifs in PfNapL are also shown in a shaded box coloured grey. Disordered/missing residues are in small caps and coloured grey. Residues that could not be aligned are shown in italics. (b) Superposition of PfNapS dimer structure (orange) onto hSET (PDB code: 2E50, green) and PfNapL (PDB code: 3FS3, purple). (c) Superposition of PfNapS dimer (coloured orange) onto hSET (PDB code: 2E50, green) and yNAP-1 (PDB code: 2AYU, purple). The 'accessory domain' of yNAP-1 which is absent in others is coloured blue.