Abstract
Pigment epithelium-derived factor (PEDF) is a multifunctional secreted glycoprotein that displays broad anti-tumor activity based on dual targeting of the tumor microenvironment (anti-angiogenic action) and the tumor cells (direct anti-tumor action). Here we show that PEDF expression is high in melanocytes, but it is lost during malignant progression of human melanoma. Using a high throughput analysis of the data from microarray studies of molecular profiling of human melanoma, we found that PEDF expression is lost in highly invasive melanomas. In paired cell lines established from the same lesion but representing the high and low extremes of malignant potential, abundant PEDF expression was restricted to the poorly aggressive counterparts. We employed RNA interference to directly address the functional consequences of PEDF silencing. PEDF knock-down in poorly aggressive melanoma cell lines augmented migration, invasion and vasculogenic mimicry, which translated into an increased in vivo metastatic potential. PEDF interference also significantly enhanced the migratory and invasive capability of normal melanocytes and moderately increased their proliferative potential. Our results demonstrate that loss of PEDF enables melanoma cells to acquire an invasive phenotype and therefore, modulation of this multifunctional factor could be critical for the malignant progression of human melanoma.
Introduction
Pigment epithelium-derived factor (PEDF) is a 50-kDa secreted glycoprotein from the serin protease inhibitor (SERPIN) superfamily (Becerra, 1997). It was initially described in retinal pigment epithelial cells (Steele et al., 1993), but later found to be expressed in several cell types (Bilak et al., 2002; Bilak et al., 1999; Quan et al., 2005; Sawant et al., 2004; Uehara et al., 2004).
Several biological activities have been ascribed to PEDF, including neurotrophic and neuroprotective properties, inhibition of proliferation and induction of differentiation and apoptosis. How PEDF controls this variety of biological responses remains largely unknown; it presumably binds to cell surface receptors to trigger various signaling cascades. Specific biological response could be determined through the expression of different PEDF receptors (PEDFR). There is evidence to support the existence of at least two different PEDFRs, each respectively specific to neuronal (Alberdi et al., 1999; Aymerich et al., 2001; Bilak et al., 1999) or endothelial cells (Filleur et al., 2005; Yamagishi et al., 2004). A PEDFR recently cloned and characterized from retina shows phospholipase A2 activity upon PEDF binding (Notari et al., 2006). The signaling pathways activated by PEDF regulate a number of key transcription factors including NF-kB (Yabe et al., 2001), NFAT (Zaichuk et al., 2004) and PPARγ (Ho et al., 2007).
Although signaling cascades elicited by PEDF are poorly defined, its functional roles are well characterized in a number of tissues, under physiological and pathological conditions. The variety of cell types and functions targeted by PEDF indicate it is a critical mediator in multiple processes; its lack or deregulation likely contributes to a number of pathologies of the eye and central nervous system (Tombran-Tink and Barnstable, 2003). More recently its potential role in cancer has been investigated in several tumor types (Ek et al., 2006a; Fernandez-Garcia et al., 2007).
PEDF is a potent endogenous inhibitor of angiogenesis, which determines proper vascularization in the adult retina and other eye compartments (Bouck, 2002; Dawson et al., 1999). These anti-angiogenic properties prompted the study of PEDF anti-tumor activity. We (Fernandez-Garcia et al., 2007; Garcia et al., 2004) and others (Doll et al., 2003; Ek et al., 2006b) uncovered a complex mechanism underlying the anti-tumor effects of PEDF, which includes indirect anti-tumor action through inhibition of angiogenesis, direct inhibition of tumor cell migration, induction of apoptosis and differentiation in certain tumor cell types.
PEDF expression changes in the course of progression of different tumor types. A number of studies show inverse correlation between PEDF levels, grade and metastatic potential of prostate adenocarcinoma (Halin et al., 2004), pancreatic adenocarcinoma (Uehara et al., 2004), glioblastoma (Guan et al., 2003), hepatocellular carcinoma (Matsumoto et al., 2004) and Wilm's tumors (Abramson et al., 2003).
Here we analyze the expression of PEDF in human melanocytes and its regulation during their malignant conversion to melanoma. Defining melanoma progression is a difficult endeavor. It has evolved from the unidirectional transition models based on pathological criteria defining the radial growth phase (RGP), vertical growth phase (VGP) and metastatic phase (M) of melanoma (Miller and Mihm, 2006) to the recently proposed reversible transition models based on the concept of molecular plasticity and reprogramming (Hendrix et al., 2007). Molecular profiling studies strongly support this later model where reversible changes between proliferative and invasive states, defined by molecular signatures, are at the core of melanoma progression (Hoek et al., 2008; Hoek et al., 2006). In this model, invasion from the growing primary tumor requires a transition from a proliferative to an invasive phenotype, which includes a number of changes enabling intravasation, survival in the circulation and extravasation at a distant site. In turn, reverse reprogramming from invasive to proliferative phenotype drives tumorigenesis at a distant site.
We have previously described the anti-tumor and anti-metastatic effects achieved by retroviral transduction of PEDF in the aggressive human melanomas A375 and UCD-Mel-N (Garcia et al., 2004). Here we use a variety of approaches to analyze the changes in PEDF expression and the functional consequences of PEDF down-regulation during malignant progression of human melanoma.
Results
PEDF is produced at high levels by skin melanocytes
Using primary cell cultures, we found that melanocytes and fibroblasts are the two cell types that express and secrete higher levels of PEDF in the skin. We detected similar high levels of PEDF by Western blot and ELISA in conditioned medium (CM) from melanocytes and skin fibroblasts but not from keratinocytes and microvascular endothelial cells (Figures 1a and 1b). These results were consistent at mRNA level measured by quantitative RT-PCR; which showed that melanocytes expressed 476- and 126-fold more PEDF mRNA than endothelial cells or keratinocytes, respectively (Figure 1c). Retinoblastoma cell line Y79 (Seigel et al., 1994), the hepatocarcinoma cell line HepG2 and primary hepatocytes (Sawant et al., 2004) were used as positive controls, showing that melanocytes expressed comparatively high levels of PEDF.
Figure 1.
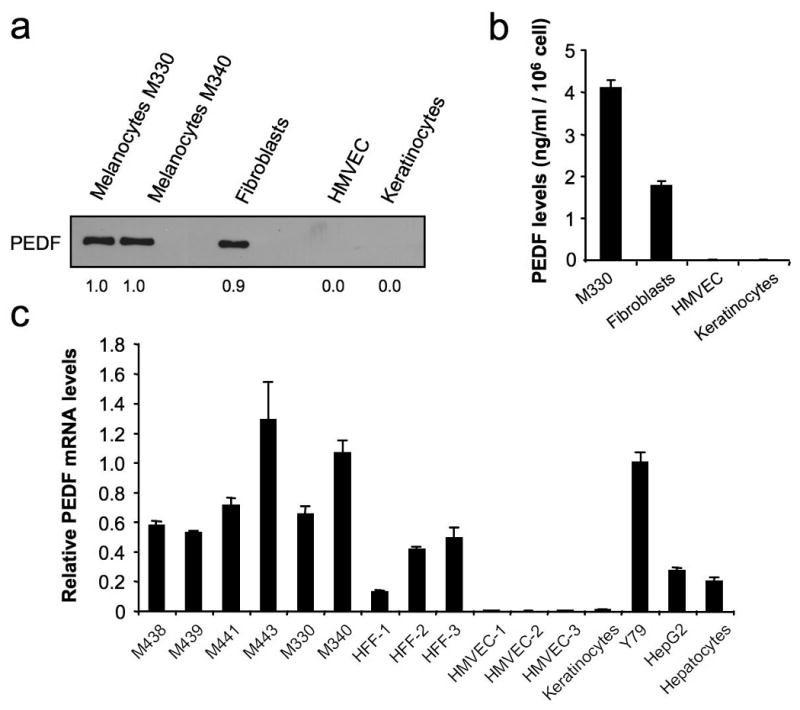
PEDF expression in primary cultures of skin cell types. (a) Western blot analysis of PEDF protein levels in 48 h-conditioned media (CM) from human primary cultures of melanocytes (M330 and M340), fibroblasts, human microvascular endothelial cells (HMVEC) and keratinocytes. Numbers below blot show densitometry values normalized to melanocytes expression. (b) ELISA analysis of secreted PEDF protein levels in 24 h-CM from human primary cultures of melanocytes (M330), fibroblasts, HMVEC and keratinocytes. Bars represent average ± standard deviation (SD). (c) Quantitative RT-PCR analysis of PEDF mRNA levels in human primary cultures of melanocytes (M438, M439, M441, M443, M330, M340), fibroblasts (HFF1, HFF2, HFF3), endothelial cells (HMVEC1, HMVEC2, HMVEC3), keratinocytes, Y79 retinoblastoma cell line, HepG2 hepatocarcinoma cell line and hepatocytes. PEDF mRNA levels are shown relative to Y79 after normalization to 18S rRNA. Bars represent average ± SD.
PEDF expression is lost in highly invasive melanomas
To determine whether PEDF expression was modulated during melanoma genesis and progression, we measured PEDF expression in a small series of human melanoma cell lines representative of the radial growth phase (RGP), vertical growth phase (VGP) and metastatic phase (M) of human melanoma. Western blot of CM showed that some RGP as well as VGP cell lines continue to express PEDF at levels similar to the primary cultures of melanocytes; however, nearly all metastatic cell lines tested lacked PEDF expression (Figure 2a). This was further confirmed by ELISA (Figure 2b) and correlated with levels of intracellular PEDF protein (Figure 2c) and PEDF mRNA levels measured by quantitative RT-PCR (Figure 2d). Mean PEDF mRNA level determined in primary melanocytes from 6 different donors was 24-fold higher than the mean of our collection of 10 melanoma cell lines (p<0.001). However, among the metastatic cell lines, we found some exceptions where PEDF is expressed at significant levels, like WM164 (Figure 2).
Figure 2.
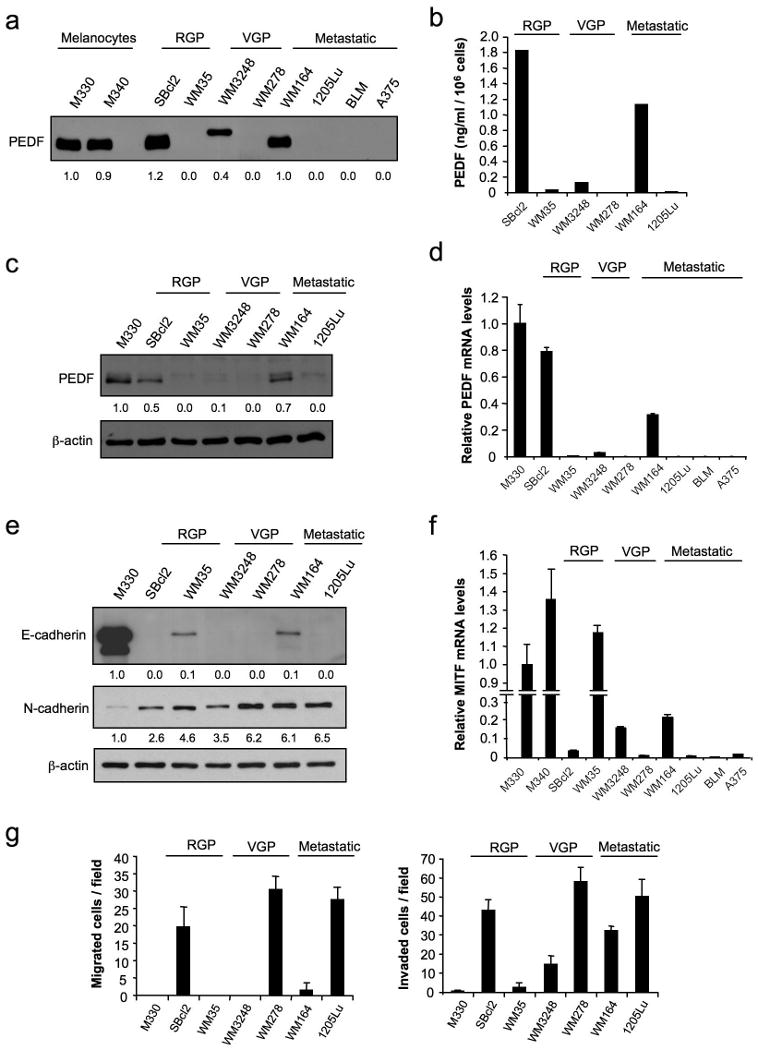
PEDF expression in RGP, VGP and metastatic human melanoma cell lines compared to normal human melanocytes. (a) Immunoblotting analysis of PEDF protein levels in 48 h-CM from M330 and M340 primary melanocytes; SBcl2 and WM35 RGP melanoma cell lines; WM3248 and WM278 VGP melanoma cell lines; and WM164, 1205Lu, BLM and A375 metastatic melanoma cell lines. Numbers below blot show densitometry values normalized to M330 melanocytes expression. (b) ELISA analysis of secreted PEDF protein levels in 48 h-CM from SBcl2, WM35, WM3248, WM278, WM164 and 1205Lu melanoma cell lines. Bars represent average ± SD. (c) Immunoblotting analysis of intracellular PEDF protein levels in whole-cell extracts from M330 primary melanocytes and SBcl2, WM35, WM3248, WM278, WM164 and 1205Lu melanoma cell lines. β-actin was used as loading control. Numbers below blot show densitometry values normalized to melanocytes expression after correction for loading. (d) Quantitative RT-PCR analysis of PEDF mRNA levels in M330 primary melanocytes and SBcl2, WM35, WM3248, WM278, WM164, 1205Lu, BLM and A375 melanoma cell lines. PEDF mRNA levels are shown relative to melanocytes after normalization to 18S rRNA. Bars represent average ± SD. (e) Immunoblotting analysis of E-cadherin and N-cadherin levels in whole-cell protein extracts from M330 primary melanocytes and SBcl2, WM35, WM3248, WM278, WM164 and 1205Lu melanoma cell lines. β-actin was used as loading control. Numbers below blots show densitometry values normalized to melanocytes expression after correction for loading. (f) Quantitative RT-PCR analysis of MITF mRNA levels in M330 primary melanocytes and SBcl2, WM35, WM3248, WM278, WM164, 1205Lu, BLM and A375 melanoma cell lines. MITF mRNA levels are shown relative to melanocytes after normalization to 18S rRNA. Bars represent average ± SD. (g) Migration (left panel) and invasion (right panel) assays of M330 primary melanocytes and SBcl2, WM35, WM3248, WM278, WM164 and 1205Lu melanoma cell lines towards 10% fetal bovine serum (FBS) for 8 h (migration) or 24 h (invasion).
To further link decreased PEDF expression and melanoma aggressiveness we characterized molecular (E-cadherin (Li et al., 2001b), N-cadherin (Li et al., 2001a) and Microphthalmia-associated transcription factor (MITF) (Carreira et al., 2006)) and functional (migration and invasion) markers of melanoma progression (Figure 2e-g). Among the VGP and metastatic cell lines tested, PEDF was expressed only in WM3248 and WM164 cell lines, which retained the expression of either E-cadherin or MITF, resulting in a lower migratory and invasive capability. However, this correlation was not observed in the two RGP cell lines selected. WM35 had lost PEDF expression but expressed E-cadherin and MITF and showed no migratory or invasive capability. Conversely, SBcl2 expressed high levels of PEDF despite its high migratory and invasive potential, which was consistent with the loss of MITF and E-cadherin.
Considering the inherent difficulty in classifying melanoma, and the variability in PEDF expression in our limited series of cell lines, we aimed to expand our analysis to larger melanoma sets. We used a high-throughput analysis of the data from microarray studies available at Gene Expression Omnibus (GEO) database, which includes large series of human melanoma cell lines. We first analyzed if PEDF expression was significantly altered between melanocyte and melanoma. We compared the normalized expression of PEDF averaged between the 28 independent primary melanocyte cultures versus expression in each melanoma data set (described in Supplementary Methods). Table 1a shows that there is a consistent and statistically significant down-regulation of PEDF in the four melanoma cell line collections.
Table 1.
Differences in PEDF expression between melanocytes and melanoma cell line collections and within melanoma cell line collections
| a. Differences in PEDF expression between melanocyte and melanoma cell line collections. | ||||||
|---|---|---|---|---|---|---|
| Data set | Average | S.D. | Fold | Statistical test | ||
| Melanocyte | 3.58 | 3.25 | ||||
| Mannheim | 1.45 | 1.98 | 2.48 | Mann-Whitney, p<0.001 | ||
| Melanocyte | 3.44 | 3.13 | ||||
| Philadelphia | 0.84 | 1.22 | 4.10 | Mann-Whitney, p<0.0001 | ||
| Melanocyte | 5.47 | 4.97 | ||||
| Johansson | 2.63 | 5.03 | 2.08 | Mann-Whitney, p<0.0001 | ||
| Melanocyte | 2.27 | 2.06 | ||||
| Wagner | 0.61 | 0.82 | 3.75 | Mann-Whitney, p<0.001 | ||
| b. Differences in PEDF expression between weakly and strongly invasive cohorts of melanoma cell lines. | ||||||
| Data set | Weakly invasive | Strongly invasive | Fold | Statistical test | ||
| Average | S.D. | Average | S.D. | |||
| Mannheim | 4.74 | 4.67 | 1.05 | 1.59 | 4.50 | Mann-Whitney, p<0.05 |
| Philadelphia | 4.99 | 5.94 | 2.16 | 4.58 | 2.31 | Mann-Whitney, p<0.05 |
| Johansson | 8.21 | 15.48 | 3.11 | 6.27 | 2.64 | Mann-Whitney, not significant (p=0.14) |
| Wagner | 2.65 | 2.79 | 1.48 | 2.31 | 1.78 | t-test, not significant (p=0.42) |
Note: while “Melanocyte” refers to the same collection of cultures in each case, the normalized expression measurement is affected by the melanoma cell line data set with which it is normalized and therefore the average value for PEDF should not be expected to remain static.
The attempts to classify melanoma according to molecular profiling have introduced a number of relevant concepts. The study by Hoek and coworkers (Hoek et al., 2006) identified molecular signatures differentiating weakly vs. strongly invasive cohorts. We applied this classification to the four melanoma cell line series above, and we questioned if PEDF was differentially expressed in the two cohorts. Figure 3 shows that PEDF is expressed at lower levels in the strongly invasive cohort in all series; this decrease was statistically significant in Mannheim and Philadelphia series (Table 1b).
Figure 3.
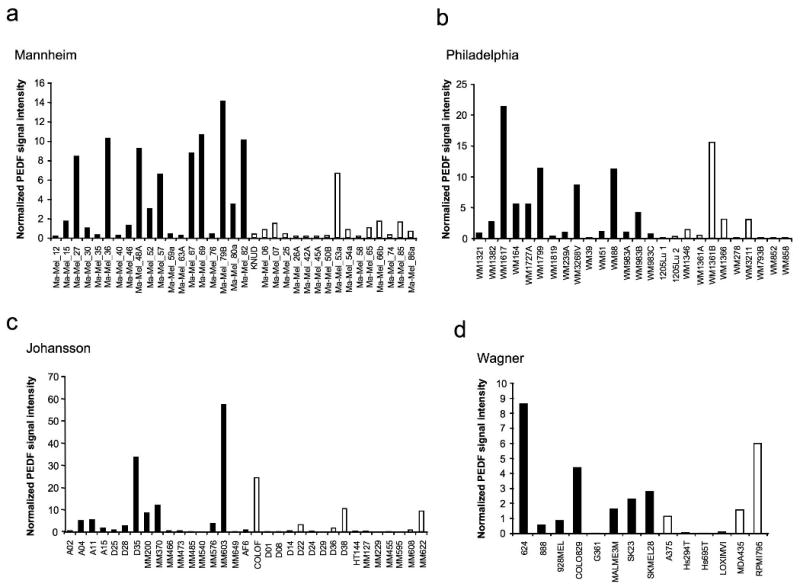
PEDF expression in series of melanoma cell lines from microarray studies. Analysis of PEDF expression utilizing microarray gene expression data of Mannheim (a), Philadelphia (b), Johansson (c) and Wagner (d) series of melanoma cell lines. Cell lines were classified by their gene signature into weakly (filled bars) and strongly invasive (empty bars) cohorts as described in Supplementary Methods. Normalized PEDF signal intensity is plotted across the sample set. Average and SD values and statistical significance are described in Table 1b.
Despite the general trend towards PEDF loss during melanoma progression, we found some metastatic melanoma cell lines that retained high PEDF levels. We then hypothesized that (i) PEDF expression is reprogrammed during the metastatic process and (ii) high PEDF expression is typical of the poorly invasive sub-population within a metastatic lesion. To verify this hypothesis, we used paired cell lines representing the extreme phenotypes (highly and poorly aggressive) derived from an abdominal wall metastasis of a cutaneous human melanoma (Seftor et al., 2005). Figure 4a shows morphological differences between the two cell lines, spindle-shaped poorly aggressive C81-61 and epithelioid highly aggressive C8161. Both C8161 and C81-61 expressed N-cadherin but have lost E-cadherin expression (Figure 4b); however their aggressive traits differed significantly. C81-61 was less migratory (Figure 4c left) and invasive (Figure 4c right), and grew poorly in soft agar (Figure 4d). Accordingly, MITF expression was higher in the poorly aggressive C81-61 than in highly aggressive C8161 (Figure 4e). Analysis of PEDF expression revealed a striking difference between the two cell lines: poorly aggressive C81-61 expressed PEDF at levels comparable to those in melanocytes, whereas PEDF was undetectable in highly aggressive C8161, as was shown by Western blot (Figure 4f left) and ELISA (Figure 4f middle) of CM and Western blot of cell extracts (Figure 4f right). Similarly, PEDF mRNA was high in C81-61 but undetectable in C8161 (Figure 4g).
Figure 4.
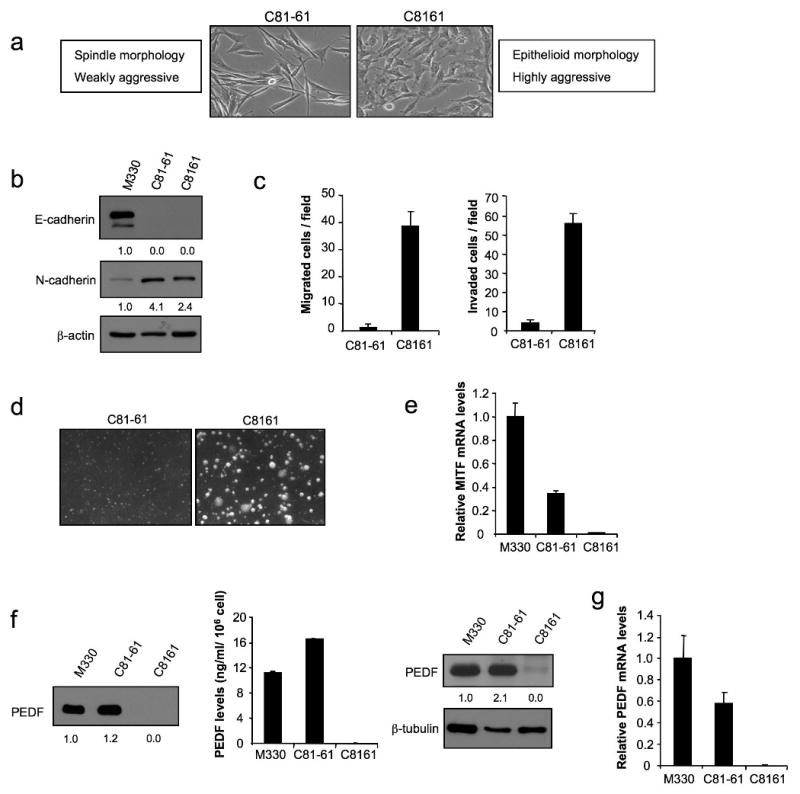
PEDF expression in melanoma cell lines representing extreme phenotypes (poorly and highly aggressive) derived from an abdominal wall metastasis of a cutaneous human melanoma. (a) Phase-contrast images of C81-61 (poorly aggressive) and C8161 (highly aggressive) melanoma cell lines. (b) Immunoblotting analysis of E-cadherin and N-cadherin levels in whole-cell protein extracts from M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. β-actin was used as loading control. Numbers below blots show densitometry values normalized to melanocytes expression after correction for loading. (c) Migration (left panel) and invasion (right panel) assays of C81-61 and C8161 melanoma cell lines towards 10% FBS for 8 h (migration) or 24 h (invasion). (d) Phase-contrast images of C81-61 and C8161 melanoma cell lines growing in soft agar after 11 days of culture. (e) Quantitative RT-PCR analysis of MITF mRNA levels in M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. MITF mRNA levels are shown relative to melanocytes after normalization to 18S rRNA. Bars represent average ± SD. (f) Immunoblotting analysis of PEDF protein levels in 48 h-CM from M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. Numbers below blot show densitometry values normalized to melanocytes expression (left panel). ELISA analysis of secreted PEDF protein levels in 48 h-CM from M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. Bars represent average ± SD (middle panel). Immunoblotting analysis of intracellular PEDF protein levels in whole-cell extracts from M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. β-tubulin was used as loading control. Numbers below blot show densitometry values normalized to melanocytes expression after correction for loading (right panel). (g) Quantitative RT-PCR analysis of PEDF mRNA levels in M330 primary melanocytes and C81-61 and C8161 melanoma cell lines. PEDF mRNA levels are shown relative to melanocytes after normalization to 18S rRNA. Bars represent average ± SD.
Heterogeneity of PEDF expression within a metastatic lesion was confirmed by immunohistochemistry in biopsies of lymph node and dermal metastasis. PEDF expression was highly positive in one out of five cases for both types of metastasis and all positive cases showed heterogeneous staining (Supplementary Figure 1).
PEDF silencing augments migratory capability, metastatic potential and vasculogenic mimicry of poorly aggressive melanoma cell lines
To establish a causal connection between PEDF expression and melanoma aggressiveness and invasiveness, we explored the functional consequences of PEDF silencing in melanoma cell lines. We knocked-down PEDF in poorly aggressive SBcl2 and WM164 melanoma cell lines using shRNAmir to PEDF (shPEDF) delivered by lentiviral transduction (Figure 5a). Non-silencing shRNAmir (NS) was used as control. PEDF silencing was quantified by Western blot (Figure 5b) and ELISA (Figure 5c) from CM and quantitative RT-PCR (Figure 5d). Basal and induced migration was increased upon interference of PEDF in SBcl2 and WM164 cells (Figure 5e). Moreover, invasion of SBcl2 was also significantly increased by PEDF silencing (Figure 5f). To evaluate if the increase in migratory ability translated into augmented metastatic potential, we tested the capacity of SBcl2-NS vs. SBcl2-shPEDF to form metastatic colonies in vivo. Interestingly, lungs of SBcl2-shPEDF-injected mice showed a higher number of microscopic GFP-positive tumor colonies compared to the lungs from SBcl2-NS-injected mice (Figure 5g, upper panels). Histological examination confirmed the presence of larger metastases in SBcl2-shPEDF colonized lungs (Figure 5g, lower panels). Quantitative analysis revealed a significant increase in the number of colonies due to PEDF silencing (Figure 5g, right).
Figure 5.
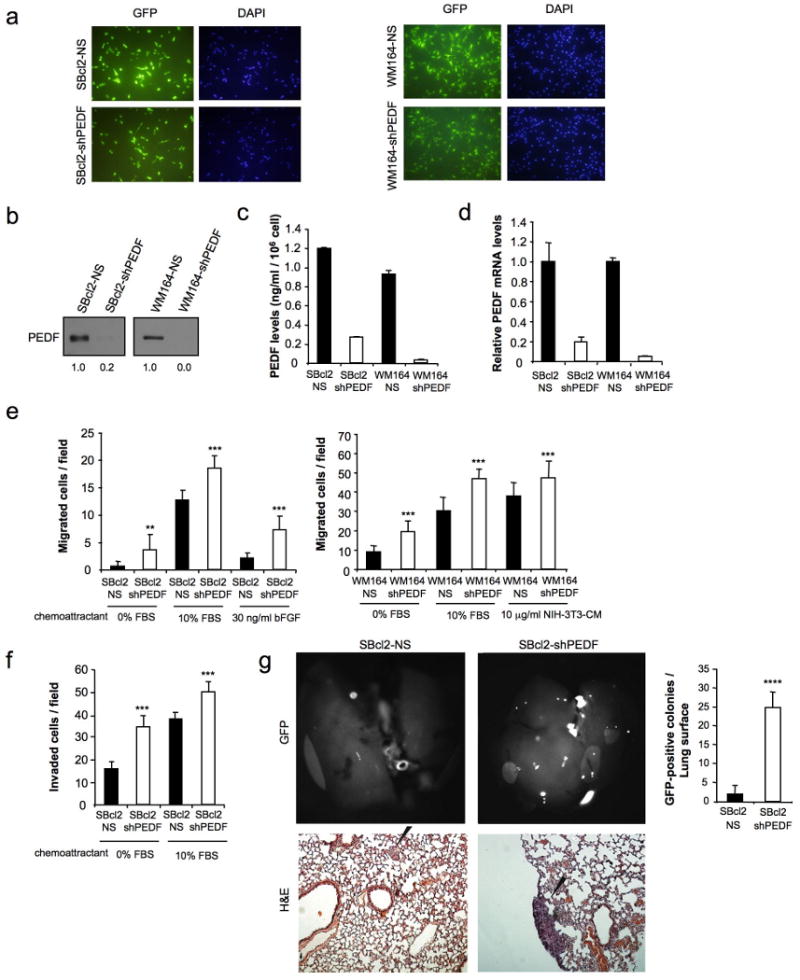
PEDF interference in poorly aggressive melanoma cell lines increases cell migration, invasion and metastatic potential. (a) Transduction efficiency of SBcl2 (left panels) and WM164 (right panels) melanoma cell lines after infection with non-silencing (NS) or shRNAmir to PEDF (shPEDF) lentivirus at a multiplicity of infection (MOI) of 10 (SBcl2) or 60 (WM164). Fluorescence images show more than 95% GFP-positive cells. Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. (b) Immunoblotting analysis of secreted PEDF protein levels in 48 h-CM from SBcl2-NS, SBcl2-shPEDF (left), WM164-NS and WM164-shPEDF (right) melanoma cell lines. Numbers below blots show densitometry values normalized to NS cell lines expression. (c) ELISA analysis of secreted PEDF protein levels in 48 h-CM from SBcl2-NS, SBcl2-shPEDF, WM164-NS and WM164-shPEDF melanoma cell lines. Bars represent average ± SD. (d) Quantitative RT-PCR analysis of PEDF mRNA levels in SBcl2-NS, SBcl2-shPEDF, WM164-NS and WM164-shPEDF melanoma cell lines. PEDF mRNA levels are shown relative to NS cell lines after normalization to GAPDH. Bars represent average ± SD. (e) Migration assays of SBcl2-NS and SBcl2-shPEDF (left) and WM164-NS and WM164-shPEDF (right) melanoma cell lines towards 10% FBS, 30 ng/ml basic fibroblast growth factor (bFGF) or 10 μg/ml CM from NIH-3T3 cells for 8 h (SBcl2) or 16 h (WM164). Statistical significance was determined by ANOVA test using Tukey-Kramer post-test (**, p<0.01; ***, p<0.001). (f) Invasion assay of SBcl2-NS and SBcl2-shPEDF melanoma cell lines towards 10% FBS for 24 h. Statistical significance was determined by ANOVA test using Tukey-Kramer post-test (***, p<0.001). (g) Lung colonization assay of SBcl2-NS and SBcl2-shPEDF melanoma cell lines. Lung metastases were visualized by fluorescence imaging. Fluorescence images of the lungs (top, left) and quantification of surface metastases (top, right) are shown. Bars represent average ± SD, and statistical significance was determined by Student's t-test (****, p<0.0001). Bottom panels show Hematoxylin/Eosin (H&E) staining of lung sections, and microscopic metastases are pointed by black arrows.
We also examined the effect of PEDF interference in the poorly aggressive melanoma cell line C81-61. Despite effective PEDF silencing (Figure 6a), there was no significant increase in migration (data not shown). As mentioned previously, melanoma cells possess extreme plasticity and can be reprogrammed to form vessel-like structures, a phenomenon termed vasculogenic mimicry (VM) (Hendrix et al., 2003). Highly aggressive C8161 cells form vasculogenic networks, unlike the poorly aggressive C81-61 that adhere poorly to collagen matrix and fail to form cord-like structures (Seftor et al., 2005). Interestingly, PEDF knock-down enabled C81-61 cells to spread out and form circular vasculogenic structures in 3D collagen matrix, indicative of VM, while non-silencing shRNAmir had no effect (Figure 6b). C81-61-shPEDF cells were not able to form colonies in the lungs 3 weeks after tail vein injection (Figure 6c). Interestingly, PEDF knock-down provided signals that allowed the melanoma cells to form branching, vessel-like structures and large lacunae containing red blood cells (Figure 6c top). Immunostaining for GFP confirmed that these structures were formed by melanoma cells (Figure 6c bottom). However, PEDF knock-down significantly increased the ability of poorly aggressive C81-61 cells implanted subcutaneously to form spontaneous metastasis to the lung after 9 weeks (Supplementary Figure 2a)
Figure 6.
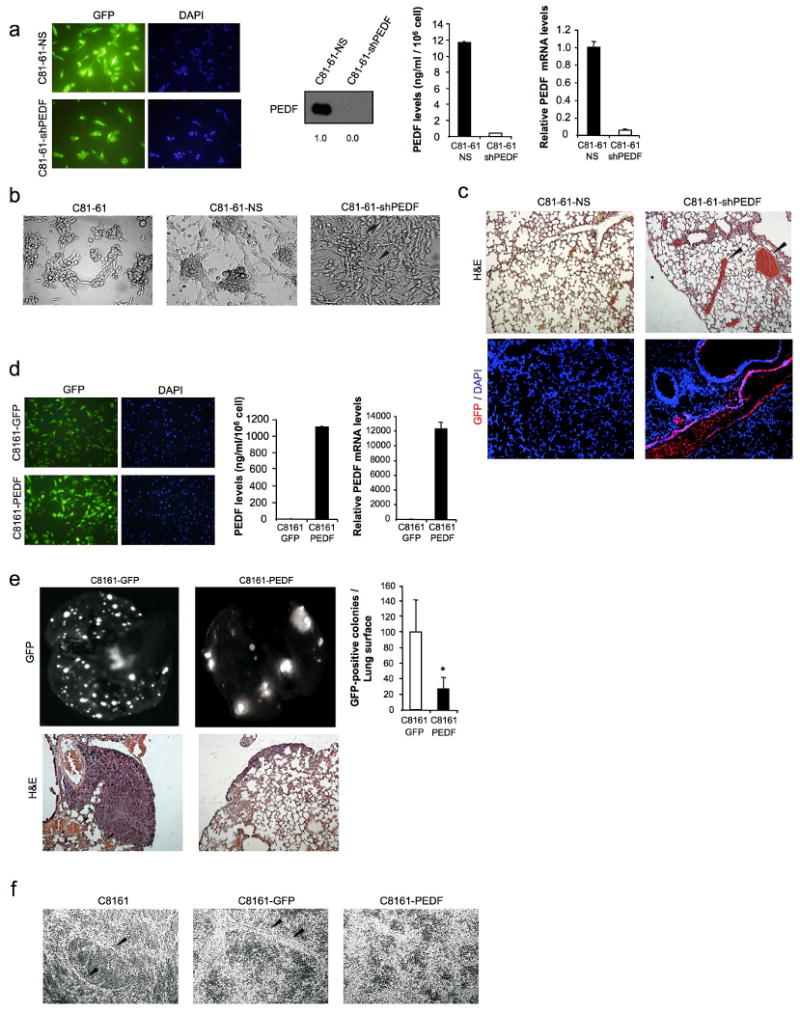
PEDF interference increases melanoma vasculogenic mimicry. (a) Transduction efficiency of C81-61 melanoma cell line after infection with non-silencing (NS) or shRNAmir to PEDF (shPEDF) lentivirus at a MOI of 40. Fluorescence images show more than 95% GFP-positive cells. Nuclei were visualized by DAPI staining (left panels). Immunoblotting analysis of secreted PEDF protein levels in 48 h-CM from C81-61-NS and C81-61-shPEDF melanoma cell lines. Numbers below blot show densitometry values normalized to C81-61-NS expression (middle left panel). ELISA analysis of secreted PEDF protein levels in 48 h-CM from C81-61-NS and C81-61-shPEDF melanoma cell lines. Bars represent average ± SD (middle right panel). Quantitative RT-PCR analysis of PEDF mRNA levels in C81-61-NS and C81-61-shPEDF melanoma cell lines. PEDF mRNA levels are shown relative to C81-61-NS after normalization to GAPDH. Bars represent average ± SD (right panel). (b) In vitro vasculogenic mimicry assay of C81-61, C81-61-NS and C81-61-shPEDF melanoma cell lines. Black arrows point to cord-like structures suggestive of vasculogenic mimicry in C81-61-shPEDF cells. (c) Formation of vasculogenic-like networks in vivo by C81-61-shPEDF melanoma cells. H&E staining of lung sections from C81-61-NS- and C81-61-shPEDF-injected mice. Vessel-like structures and blood-filled lacunae (pointed by arrows) were detected in lungs from mice inoculated with C81-61-shPEDF cells (top). Immunofluorescence staining for GFP (red) and DAPI (blue) counterstaining are shown (bottom). (d) Transduction efficiency of C8161 melanoma cell line after infection with control (GFP) or PEDF-over-expressing (PEDF) lentivirus at a MOI of 10. Fluorescence images show more than 95% GFP-positive cells. Nuclei were visualized by DAPI staining (left panels). ELISA analysis of secreted PEDF protein levels in 48 h-CM from C8161-GFP and C8161-PEDF melanoma cell lines. Bars represent average ± SD (middle panel). Quantitative RT-PCR analysis of PEDF mRNA levels in C8161-GFP and C8161-PEDF melanoma cell lines. PEDF mRNA levels are shown relative to C8161-GFP after normalization to GAPDH. Bars represent average ± SD (right panel). (e) Lung colonization assay of C8161-GFP and C8161-PEDF melanoma cell lines. Lung metastases were visualized by fluorescence imaging. Fluorescence images of the lungs (top, left) and quantification of surface metastases (top, right) are shown. Bars represent average ± SD, and statistical significance was determined by Student's t-test (*, p<0.05). Bottom panels show H&E staining of lung sections. Vessel-like structures in lungs of mice inoculated with C8161-GFP cells are indicated by arrows. (f) In vitro vasculogenic mimicry assay of C8161, C8161-GFP and C8161-PEDF melanoma cell lines. Cord-like structures in C8161 and C8161-GFP cell cultures are indicated by arrows.
We also over-expressed PEDF in the highly aggressive and invasive counterpart of C81-61 melanoma cell line, C8161. Efficient PEDF lentiviral transduction of C8161 cells was confirmed by GFP expression (Figure 6d, left) and analysis of PEDF expression by ELISA of CM (Figure 6d, middle) and quantitative RT-PCR (Figure 6d, right). In agreement with our previous observations (Garcia et al., 2004), PEDF over-expression reduced the number of surface lung metastases in a lung colonization assay (Figure 6e) and the ability to form spontaneous metastases from a subcutaneous primary tumor (Supplementary Figure 2b). Moreover, it reduced the formation of circular cords in 3D collagen matrix in vitro (Figure 6f), as well as the presence of blood-filled lacunae in vivo (Figure 6e, lower panels), indicative of VM, typical of C8161 cells (Hendrix et al., 2003; van der Schaft et al., 2004).
PEDF silencing augments migration, invasion and proliferation of melanocytes
Since PEDF is highly expressed in melanocytes, we explored if PEDF could have a direct functional role in these cells regulating their ability to migrate, invade and proliferate. PEDF knock-down in melanocytes (Figures 7a-7c) increased migration and invasion towards a variety of inducers (Figures 7d and 7e). Furthermore, the proliferation of melanocytes was moderately increased by PEDF knock-down (Figure 7f).
Figure 7.
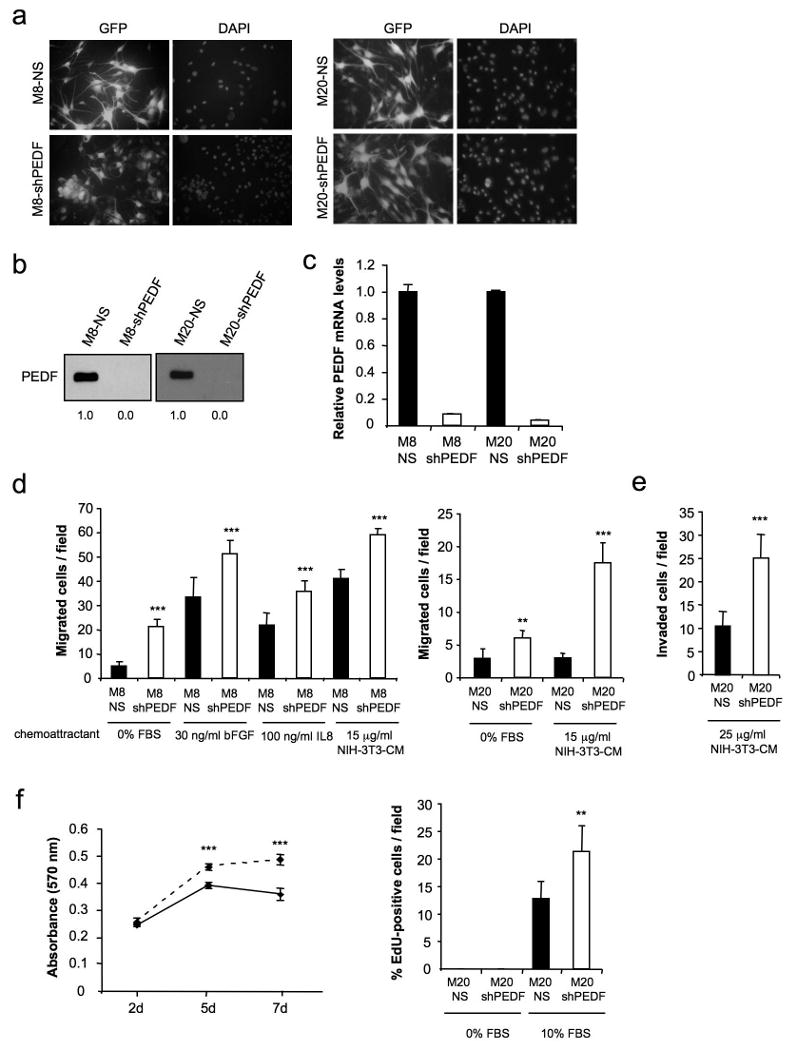
PEDF interference augments migration, invasion and proliferation of the primary melanocytes. (a) Transduction efficiency of M8 (left panels) and M20 (right panels) primary melanocytes after infection with non-silencing (NS) or shRNAmir to PEDF (shPEDF) lentivirus at a MOI of 40. Fluorescence images show more than 95% GFP-positive cells. Nuclei were visualized by DAPI staining. (b) Immunoblotting analysis of secreted PEDF protein levels in 48 h-CM from M8-NS, M8-shPEDF (left), M20-NS and M20-shPEDF (right) melanocytes. Numbers below blots show densitometry values normalized to NS cells expression. (c) Quantitative RT-PCR analysis of PEDF mRNA levels in M8-NS, M8-shPEDF, M20-NS and M20-shPEDF melanocytes. PEDF mRNA levels are shown relative to NS cells after normalization to 18S rRNA. Bars represent average ± SD. (d) Migration assay of M8-NS and M8-shPEDF (left) and M20-NS and M20-shPEDF (right) melanocytes towards 30 ng/ml bFGF, 100 ng/ml IL8 or 15 μg/ml CM from NIH-3T3 cells for 22 h. Statistical significance was determined by ANOVA using Tukey-Kramer post-test (**, p<0.01; ***, p<0.001). (e) Invasion assay of M20-NS and M20-shPEDF melanocytes towards 25 μg/ml CM from NIH-3T3 cells for 24 h. Statistical significance was determined by Student's t-test (***, p<0.001). (f) Proliferation assays. Proliferation curves of M20-NS (solid line) and M20-shPEDF (dashed line) melanocytes growing in the presence of serum. MTT assay was performed at the indicated time points (left panel). Right panel shows 5-ethynyl-2-deoxyuridine (EdU) incorporation of M20-NS and M20-shPEDF melanocytes. Cells were serum-starved for 24 h and then were allowed to grow in the presence of 20 μM EdU in serum-free medium or 10% FBS-containing medium for another 24 h. Bars represent average ± SD. Statistical significance was determined by Student's t-test (**, p<0.01; ***, p<0.01)
PEDF silencing did not alter the levels of E-cadherin protein and mRNA (Supplementary Figure 3). Although we found a reduction in MITF mRNA expression upon PEDF knock-down (Supplementary Figure 4), this result was not reproduced in a statistically significant number of experiments; MITF levels were reduced in three out of nine independent experiments using primary cultures from different donors. A reduction in MITF mRNA after PEDF silencing was also seen in the poorly aggressive melanoma cell lines WM164, C81-61 and M010817, but did not reproduce in a significant number of experiments (Supplementary Figure 4). Thus, it is unlikely that the functional effects of PEDF silencing in melanocytes were mediated by E-Cadherin or MITF.
Discussion
Our understanding of melanoma biology remains incomplete despite several decades of intense research (Gray-Schopfer et al., 2007). The role of epigenetic factors and tumor microenvironment as major driving forces in melanoma progression have been presented with increasing molecular detail in recent years (Hendrix et al., 2007).
Here we describe the regulation of PEDF, a multifunctional endogenous factor, during melanoma progression and its functional consequences. We have previously demonstrated that PEDF over-expression has multiple biological effects that overall profoundly affect melanoma aggressiveness by abrogating migration, driving apoptosis in melanoma cells under stress conditions, and acting as a potent anti-angiogenic factor (Garcia et al., 2004). These results were extrapolated to other tumor types by a number of subsequent studies, which strongly support the potential use of PEDF for cancer therapy.
Here we show that PEDF is produced at high levels by melanocytes in the skin. Originally PEDF was found at high levels in retinal pigment epithelial cells (RPE). Although both melanocytes and RPE cells are pigment producing cells, they have different developmental origin and display distinct biological roles (Sato and Yamamoto, 2001). PEDF has multiple functions, some of which are cell type-specific, like neurotrophic or neuroprotective properties, while others, including inhibition of proliferation, migration and induction of apoptosis affect multiple cell types. The functional role of PEDF in pigment producing cells has not yet been addressed. Here we show that knock-down of endogenous PEDF expression in melanocytes translates into increased migration, invasion and proliferation, suggesting a role for PEDF in melanocyte homeostasis. Further studies comparing PEDF expression in highly migratory neural crest precursors and in less migratory mature melanocytes in the epidermis, may provide valuable clues into the functional roles of PEDF in pigment producing cells.
PEDF is expressed at high levels in a significant percentage of melanoma lesions although the high variance found in its expression levels suggested that regulation of PEDF expression may underlie some aspects of melanoma progression. In our study we used several approaches to establish a robust conclusion concerning PEDF loss during melanoma progression and its functional consequences.
We utilized the information from high-throughput microarray analysis available at GEO databases to analyze possible correlation of PEDF expression with molecular staging of melanoma. The comparison of normalized data from several independent studies showed that PEDF was significantly down-regulated in melanoma cell lines compared to melanocyte cultures. This analysis also showed that PEDF expression was highly variable between melanoma cell lines. We therefore looked for a sub-classification that would reflect the principle underlying PEDF variation. Gene expression profiling has been used to define molecular taxonomy of human melanoma, which reflects certain functional criteria. Hoek and co-workers used the combination of gene expression profiling and functional tests to introduce a hypothesis in which melanoma cells are classified into weakly or strongly invasive, and bidirectional switch between these phenotypes drives progression (Hoek et al., 2008). We found that high PEDF expression was characteristic of weakly invasive melanoma cell lines.
On the other hand, our study suggested that PEDF expression in melanoma cells is subject to plasticity. This was supported by our results in paired cell lines established from a single lesion, where PEDF expression was restricted to the poorly aggressive counterpart. Therefore a single melanoma lesion may contain PEDF positive and negative cells and reprogramming of PEDF expression may reflect the reversible transitions between proliferative and invasive states required during the metastatic process. In keeping with this hypothesis, melanoma biopsies from the lymph node and dermal metastases showed heterogeneous expression of PEDF and in one out of five cases PEDF was expressed at high levels.
To assess the functional consequences of PEDF loss during malignant progression of human melanoma we used a direct molecular analysis based on RNA interference. Silencing of endogenous PEDF expression in poorly aggressive human melanoma cell lines increased their migratory and invasive ability and translated into a higher metastatic potential in vivo. Additional traits modulated by PEDF, like the ability to proliferate and induce angiogenesis (Garcia et al., 2004), could also contribute to the observed increased metastatic potential.
Interestingly, PEDF silencing increased VM, one of the phenomena reflecting melanoma plasticity during the invasion-metastasis cascade (Hendrix et al., 2003). The ability of PEDF to curtail VM is especially noteworthy, since it was unaffected by established angiogenesis inhibitors such as anginex, TNP-470, and endostatin (van der Schaft et al., 2004).
Possible regulatory mechanisms underlying the changes in PEDF expression during transformation and malignant progression of melanoma are presently unknown, although there are several likely candidates. Regulators of neovascularisation, like PEDF, are controlled through the gain of oncogenes and the loss of tumor suppressor genes (Bouck, 1990). PEDF is a direct target of p53 family members in colorectal cancer cell lines (Sasaki et al., 2005). Decreased PEDF expression correlates with the expression of ΔEX2p73, or with K-ras oncogenic mutations in human colorectal cancer (Diaz et al., 2008). The role of p73 in melanoma genesis is poorly defined. A recent study demonstrates that TA-p73 abrogates anchorage-independent growth through regulation of KCNK1, which is frequently lost in melanoma (Beitzinger et al., 2008). N-ras oncogenic mutations are found in 20% of human melanomas and provide a proliferative advantage early in transformation (Chin, 2003). Analysis of GEO data-bases for possible correlation between PEDF expression and B-RAF or N-ras mutations in melanoma yielded no significant correlation. Regulatory signals from the melanoma microenvironment may also influence PEDF expression. We found a 9-fold decrease of PEDF expression in primary melanocytes grown on collagen matrix preconditioned by a highly aggressive melanoma cell line, as compared to melanocytes grown on collagen without pre-conditioning (data not shown) (Seftor et al., 2005). Additionally, hypoxia is a relevant trigger of the angiogenic switch and other epigenetic changes contributing to metastasis. PEDF is down-regulated by hypoxia in retinoblastoma (Dawson et al., 1999) and RPE cells (Notari et al., 2005). We also found that PEDF is down-regulated by hypoxia in melanocytes and melanoma cell lines (data not shown).
In summary, our study shows that PEDF is expressed at high levels in the melanocyte, where it contributes to restrict the growth, migration and invasion of melanocytes. PEDF expression is modulated in a complex fashion during malignant progression of human melanoma, where it is likely reprogrammed to meet the current demand for proliferation, migration, invasion or angiogenesis.
Materials and methods
Cell culture
See Supplementary Methods for a detailed description of the cell lines used.
Immunoblotting
Whole-cell lysates or CM were prepared and analyzed as described (Fernandez-Garcia et al., 2005), and 15 μg whole-cell protein extracts or 1.5 μg CM were loaded per lane. Specific primary antibodies and dilutions are listed in the Supplementary Table 1.
ELISA
Secreted PEDF protein levels in CM were quantified using ELISAquant PEDF Sandwich ELISA Antigen Detection Kit following the manufacturer's instructions (Bioproducts MD LLC, MD, USA).
Immunofluorescence and immunohistochemistry
Immunofluorescence and immunohistochemistry methods are included in Supplementary Methods.
RNA extraction and quantitative RT-PCR
Total RNA was isolated using TRIzol (Molecular Research Center Inc., Cincinnati, OH, USA) and was retrotranscribed to cDNA using High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). See Supplementary Methods for the probes and oligonucleotides used. The quantitative PCR reaction was performed in an ABI Prism 7900 HT thermal cycler (Applied Biosystems) following the manufacturer's protocols. Relative mRNA levels were calculated using the comparative CT method as recommended by Applied Biosystems.
Microarray data analysis
Previously published DNA microarray data sets comprising series of primary melanocytes or melanoma cell lines were downloaded from public database websites and analyzed as indicated in Supplementary Methods.
RNA interference
PEDF knock-down was achieved using the lentiviral vector pGIPz containing the shRNAmir sequence VL2HS_221662 from Open Biosystems (Huntsville, AL, USA). Non-silencing shRNAmir sequence cloned into pGIPz vector and with no homology to known mammalian genes was used as control (Open Biosystems).
PEDF over-expression
For PEDF over-expression, we generated lentiviral constructs in prrl.CMV.EGFP.wpre.SIN vector encoding the full-length human PEDF cDNA as indicated in Supplementary Methods.
Lentivirus production and transduction of target cell lines
Lentiviruses were produced as previously described (Punzon et al., 2004). For transduction of target cells, lentiviruses at a multiplicity of infection (MOI) of 10-60 were added to the culture medium in the presence of 8 μg/ml polybrene (Sigma, St. Louis, MO, USA) for 8 h, typically resulting in more than 95% transduced (GFP-positive) cells. After 72 h, PEDF knockdown or over-expression was assessed. Then, PEDF-over-expressing cell lines were enriched for GFP expression by fluorescence-activated cell sorting. For PEDF-knock-down cell lines, cells were selected with 0.25-1 μg/ml puromycin (Calbiochem-Novabiochem Corp., La Jolla, CA, USA) for 4 days.
Migration and invasion assays
Modified Boyden chambers with polycarbonate filters (6.5 mm diameter, 8.0 μm pore size) (Corning Incorporated, Corning, NY, USA) were used to assess cell migration through 0.5% gelatin coated filters as described (Garcia et al., 2004). To evaluate cell invasion, the filters were coated with 12 μg Matrigel (primary melanocytes) or Growth Factor Reduced Matrigel (melanoma cell lines) (BD Biosciences) diluted in 100 μl serum-free medium, and air dried overnight. As chemoattractants, 10% fetal bovine serum (FBS); 30 ng/ml bFGF (Peprotech, London, UK); 100 ng/ml IL8 (Preprotech); or 10-25 μg/ml CM from NIH-3T3 cells were used. Following incubation for the indicated time periods, non-migrated cells were wiped off using a cotton swab, and the filters were stained with Diff Quik (Dade Behring, Newark, DE, USA). Migrated cells were counted at ten areas of maximum migration under a light microscope at 40× magnification. Average ± standard deviation (SD) values shown are representative of at least three independent experiments.
Proliferation assays
Melanocyte S phase entry was analyzed by the incorporation of 5-ethynyl-2-deoxyuridine (EdU) using the Click-iT EdU Imaging Kit (Invitrogen) as indicated by the manufacturer. EdU-positive cells were counted in six different fields using a TCS SP5 DMI6000 spectral confocal microscope (Leica Microsystems). Melanocyte viability was also measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma), following the manufacturer's instructions. Results using EdU and MTT were confirmed in two independent primary cultures, and a representative experiment is shown.
In vitro vasculogenic mimicry assay
Tissue culture plates were coated with collagen I (BD Discovery Labware), soaked in ethanol, and rinsed in PBS. 5×105 cells were seeded on the matrix in 24-well plates in regular growth media and left to grow for up to 8 days. Images were taken on day 6.
Colony formation assay
2.5×104 cells were resuspended in 0.35% Noble agar (BD Biosciences, Bedford, MA, USA) in RPMI with 10% FBS and seeded on 0.5% Noble agar in the same medium in 6-well plates. Phase-contrast micrographs were taken after 11 days of culture using a magnifying glass at × 0.7.
Lung colonization assay
NOD/SCID mice (Harlan Laboratories, Madison, WI, USA; 5 mice per group) were injected via the tail vein with melanoma cells suspended in 100 μl PBS (1×106 cells for SBcl2-NS, SBcl2-shPEDF, C81-61-NS and C81-61-shPEDF cell lines; 1×105 cells for C8161-GFP or C8161-PEDF cell lines). At the endpoint (92 days for SBcl2-NS- and SBcl2-shPEDF-injected mice; 21 days for C81-61-NS- and C81-61-shPEDF-injected mice; and 28 days for C8161-GFP- and C8161-PEDF-injected mice) the mice were sacrificed, the lungs removed and photographed using Olympus OV100 Small Animal Imaging System, and surface metastases were counted manually. The lungs were formalin-fixed, paraffin embedded and sectioned for further analysis. H&E staining was performed at Northwestern University Pathology Core.
Spontaneous metastasis assay
Melanoma cells were injected subcutaneously into the right flank of nude mice (Harlan Laboratories) (1×106 cells/flank for C8161-derived cell lines and 2×106 cells/flank for C81-61-derived cell lines; 10 mice/cell line) and the tumors were allowed to grow for 4 weeks (C8161-derived cell lines) or 9 weeks (C81-61-derived cell lines). The animals were sacrificed, the lungs and livers excised, rinsed in PBS and fluorescence images were taken using Olympus OV100 Small Animal Imaging System. The number of surface lesions was determined on digital images using MetaMorph software package.
Statistical analysis
All statistical analyses were performed using GraphPad Instat (GraphPad Software, San Diego, CA, USA). P values ≤ 0.05 were considered as significant.
Supplementary Material
Acknowledgments
We acknowledge with gratitude all researchers that contributed with cell lines: M Herlyn, PF Peñas, F Vidal-Vanaclocha, GN Van Muijen, M Garcia and M del Rio. We also thank P Fernández for her technical assistance in lentivirus production.
Supported by grants: Ministerio de Educación y Ciencia grant SAF2007-62292 (BJ), Comunidad de Madrid SAL-0311-2006 (BJ), NIH grant RO1 HL68033 (OV), NIH merit grant CA59702 (MJCH). JL Orgaz has been supported by a Ministerio de Educación y Ciencia fellowship, O Ladhani by a NIH/NCI training grant T32CA009560 and A Fernández-Barral by a Consejo Superior de Investigaciones Científicas fellowship.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Supplementary information is available at Oncogene's website.
References
- Abramson LP, Stellmach V, Doll JA, Cornwell M, Arensman RM, Crawford SE. Wilms' tumor growth is suppressed by antiangiogenic pigment epithelium-derived factor in a xenograft model. J Pediatr Surg. 2003;38:336–42. doi: 10.1053/jpsu.2003.50104. [DOI] [PubMed] [Google Scholar]
- Alberdi E, Aymerich MS, Becerra SP. Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J Biol Chem. 1999;274:31605–12. doi: 10.1074/jbc.274.44.31605. [DOI] [PubMed] [Google Scholar]
- Aymerich MS, Alberdi EM, Martinez A, Becerra SP. Evidence for pigment epithelium-derived factor receptors in the neural retina. Invest Ophthalmol Vis Sci. 2001;42:3287–93. [PubMed] [Google Scholar]
- Becerra SP. Structure-function studies on PEDF. A noninhibitory serpin with neurotrophic activity. Adv Exp Med Biol. 1997;425:223–37. [PubMed] [Google Scholar]
- Beitzinger M, Hofmann L, Oswald C, Beinoraviciute-Kellner R, Sauer M, Griesmann H, et al. p73 poses a barrier to malignant transformation by limiting anchorage-independent growth. Embo J. 2008;27:792–803. doi: 10.1038/emboj.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilak MM, Becerra SP, Vincent AM, Moss BH, Aymerich MS, Kuncl RW. Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. J Neurosci. 2002;22:9378–86. doi: 10.1523/JNEUROSCI.22-21-09378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilak MM, Corse AM, Bilak SR, Lehar M, Tombran-Tink J, Kuncl RW. Pigment epithelium-derived factor (PEDF) protects motor neurons from chronic glutamate-mediated neurodegeneration. J Neuropathol Exp Neurol. 1999;58:719–28. doi: 10.1097/00005072-199907000-00006. [DOI] [PubMed] [Google Scholar]
- Bouck N. Tumor angiogenesis: the role of oncogenes and tumor suppressor genes. Cancer Cells. 1990;2:179–85. [PubMed] [Google Scholar]
- Bouck N. PEDF: anti-angiogenic guardian of ocular function. Trends Mol Med. 2002;8:330–4. doi: 10.1016/s1471-4914(02)02362-6. [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–39. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3:559–70. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, et al. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–8. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- Diaz R, Pena C, Silva J, Lorenzo Y, Garcia V, Garcia JM, et al. p73 Isoforms affect VEGF, VEGF165b and PEDF expression in human colorectal tumors: VEGF165b downregulation as a marker of poor prognosis. Int J Cancer. 2008;123:1060–7. doi: 10.1002/ijc.23619. [DOI] [PubMed] [Google Scholar]
- Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, et al. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9:774–80. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- Ek ET, Dass CR, Choong PF. PEDF: a potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol Med. 2006a;12:497–502. doi: 10.1016/j.molmed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Ek ET, Dass CR, Choong PF. Pigment epithelium-derived factor: a multimodal tumor inhibitor. Mol Cancer Ther. 2006b;5:1641–6. doi: 10.1158/1535-7163.MCT-06-0107. [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia NI, Palmer HG, Garcia M, Gonzalez-Martin A, del Rio M, Barettino D, et al. 1alpha,25-Dihydroxyvitamin D3 regulates the expression of Id1 and Id2 genes and the angiogenic phenotype of human colon carcinoma cells. Oncogene. 2005;24:6533–44. doi: 10.1038/sj.onc.1208801. [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia NI, Volpert OV, Jimenez B. Pigment epithelium-derived factor as a multifunctional antitumor factor. J Mol Med. 2007;85:15–22. doi: 10.1007/s00109-006-0111-z. [DOI] [PubMed] [Google Scholar]
- Filleur S, Volz K, Nelius T, Mirochnik Y, Huang H, Zaichuk TA, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–52. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- Garcia M, Fernandez-Garcia NI, Rivas V, Carretero M, Escamez MJ, Gonzalez-Martin A, et al. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–42. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Guan M, Yam HF, Su B, Chan KP, Pang CP, Liu WW, et al. Loss of pigment epithelium derived factor expression in glioma progression. J Clin Pathol. 2003;56:277–82. doi: 10.1136/jcp.56.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halin S, Wikstrom P, Rudolfsson SH, Stattin P, Doll JA, Crawford SE, et al. Decreased pigment epithelium-derived factor is associated with metastatic phenotype in human and rat prostate tumors. Cancer Res. 2004;64:5664–71. doi: 10.1158/0008-5472.CAN-04-0835. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3:411–21. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7:246–55. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- Ho TC, Chen SL, Yang YC, Liao CL, Cheng HC, Tsao YP. PEDF induces p53-mediated apoptosis through PPAR gamma signaling in human umbilical vein endothelial cells. Cardiovasc Res. 2007;76:213–23. doi: 10.1016/j.cardiores.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–6. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001a;61:3819–25. [PubMed] [Google Scholar]
- Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene. 2001b;20:8125–35. doi: 10.1038/sj.onc.1205034. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Ishikawa H, Nishimura D, Hamasaki K, Nakao K, Eguchi K. Antiangiogenic property of pigment epithelium-derived factor in hepatocellular carcinoma. Hepatology. 2004;40:252–9. doi: 10.1002/hep.20259. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–37. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- Notari L, Miller A, Martinez A, Amaral J, Ju M, Robinson G, et al. Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Invest Ophthalmol Vis Sci. 2005;46:2736–47. doi: 10.1167/iovs.04-1489. [DOI] [PubMed] [Google Scholar]
- Punzon I, Criado LM, Serrano A, Serrano F, Bernad A. Highly efficient lentiviral-mediated human cytokine transgenesis on the NOD/scid background. Blood. 2004;103:580–2. doi: 10.1182/blood-2003-07-2298. [DOI] [PubMed] [Google Scholar]
- Quan GM, Ojaimi J, Li Y, Kartsogiannis V, Zhou H, Choong PF. Localization of pigment epithelium-derived factor in growing mouse bone. Calcif Tissue Int. 2005;76:146–53. doi: 10.1007/s00223-004-0068-2. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Naishiro Y, Oshima Y, Imai K, Nakamura Y, Tokino T. Identification of pigment epithelium-derived factor as a direct target of the p53 family member genes. Oncogene. 2005;24:5131–6. doi: 10.1038/sj.onc.1208695. [DOI] [PubMed] [Google Scholar]
- Sato S, Yamamoto H. Development of pigment cells in the brain of ascidian tadpole larvae: insights into the origins of vertebrate pigment cells. Pigment Cell Res. 2001;14:428–36. doi: 10.1034/j.1600-0749.2001.140602.x. [DOI] [PubMed] [Google Scholar]
- Sawant S, Aparicio S, Tink AR, Lara N, Barnstable CJ, Tombran-Tink J. Regulation of factors controlling angiogenesis in liver development: a role for PEDF in the formation and maintenance of normal vasculature. Biochem Biophys Res Commun. 2004;325:408–13. doi: 10.1016/j.bbrc.2004.10.041. [DOI] [PubMed] [Google Scholar]
- Seftor EA, Brown KM, Chin L, Kirschmann DA, Wheaton WW, Protopopov A, et al. Epigenetic transdifferentiation of normal melanocytes by a metastatic melanoma microenvironment. Cancer Res. 2005;65:10164–9. doi: 10.1158/0008-5472.CAN-05-2497. [DOI] [PubMed] [Google Scholar]
- Seigel GM, Tombran-Tink J, Becerra SP, Chader GJ, Diloreto DA, Jr, del Cerro C, et al. Differentiation of Y79 retinoblastoma cells with pigment epithelial-derived factor and interphotoreceptor matrix wash: effects on tumorigenicity. Growth Factors. 1994;10:289–97. doi: 10.3109/08977199409010995. [DOI] [PubMed] [Google Scholar]
- Steele FR, Chader GJ, Johnson LV, Tombran-Tink J. Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc Natl Acad Sci U S A. 1993;90:1526–30. doi: 10.1073/pnas.90.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombran-Tink J, Barnstable CJ. PEDF: a multifaceted neurotrophic factor. Nat Rev Neurosci. 2003;4:628–36. doi: 10.1038/nrn1176. [DOI] [PubMed] [Google Scholar]
- Uehara H, Miyamoto M, Kato K, Ebihara Y, Kaneko H, Hashimoto H, et al. Expression of pigment epithelium-derived factor decreases liver metastasis and correlates with favorable prognosis for patients with ductal pancreatic adenocarcinoma. Cancer Res. 2004;64:3533–7. doi: 10.1158/0008-5472.CAN-03-3725. [DOI] [PubMed] [Google Scholar]
- van der Schaft DW, Seftor RE, Seftor EA, Hess AR, Gruman LM, Kirschmann DA, et al. Effects of angiogenesis inhibitors on vascular network formation by human endothelial and melanoma cells. J Natl Cancer Inst. 2004;96:1473–7. doi: 10.1093/jnci/djh267. [DOI] [PubMed] [Google Scholar]
- Yabe T, Wilson D, Schwartz JP. NFkappaB activation is required for the neuroprotective effects of pigment epithelium-derived factor (PEDF) on cerebellar granule neurons. J Biol Chem. 2001;276:43313–9. doi: 10.1074/jbc.M107831200. [DOI] [PubMed] [Google Scholar]
- Yamagishi S, Inagaki Y, Nakamura K, Abe R, Shimizu T, Yoshimura A, et al. Pigment epithelium-derived factor inhibits TNF-alpha-induced interleukin-6 expression in endothelial cells by suppressing NADPH oxidase-mediated reactive oxygen species generation. J Mol Cell Cardiol. 2004;37:497–506. doi: 10.1016/j.yjmcc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Zaichuk TA, Shroff EH, Emmanuel R, Filleur S, Nelius T, Volpert OV. Nuclear factor of activated T cells balances angiogenesis activation and inhibition. J Exp Med. 2004;199:1513–22. doi: 10.1084/jem.20040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


